Uncovering the salt response of soybean by unraveling its wild and cultivated functional genomes using tag sequencing
- PMID: 23209559
- PMCID: PMC3509101
- DOI: 10.1371/journal.pone.0048819
Uncovering the salt response of soybean by unraveling its wild and cultivated functional genomes using tag sequencing
Abstract
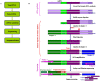
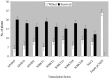
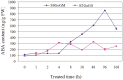
Soil salinity has very adverse effects on growth and yield of crop plants. Several salt tolerant wild accessions and cultivars are reported in soybean. Functional genomes of salt tolerant Glycine soja and a salt sensitive genotype of Glycine max were investigated to understand the mechanism of salt tolerance in soybean. For this purpose, four libraries were constructed for Tag sequencing on Illumina platform. We identify around 490 salt responsive genes which included a number of transcription factors, signaling proteins, translation factors and structural genes like transporters, multidrug resistance proteins, antiporters, chaperons, aquaporins etc. The gene expression levels and ratio of up/down-regulated genes was greater in tolerant plants. Translation related genes remained stable or showed slightly higher expression in tolerant plants under salinity stress. Further analyses of sequenced data and the annotations for gene ontology and pathways indicated that soybean adapts to salt stress through ABA biosynthesis and regulation of translation and signal transduction of structural genes. Manipulation of these pathways may mitigate the effect of salt stress thus enhancing salt tolerance.
Conflict of interest statement
Figures




References
-
- Flowers TJ, Yeo AR (1995) Breeding for salinity resistance in crop plants: where next?. Australian Journal of Plant Physiology 22: 875–884.
-
- Flowers TJ, Koyama ML, Flowers SA, Sudhakar C, Singh KP, et al. (2000) QTL: their place in engineering tolerance of rice to salinity. Journal of Experimental Botany 51: 99–106. - PubMed
-
- Cushman JC, Bohnert HJ (2000) Genomic approaches to plant stress tolerance. Current Opinion in Plant Biology 3: 117–124. - PubMed
-
- Tomy S, Chang Y-M, Chen Y-H, Cao J-C, Wang T-P, et al. (2009) Salinity effects on the expression of osmoregulatory genes in the euryhaline black porgy Acanthopagrus schlegeli . General and Comparative Endocrinology 161: 123–132. - PubMed
-
- Manaa A, Ben Ahmed H, Valot B, Bouchet JP, Aschi-Smiti S, et al. (2011) Salt and genotype impact on plant physiology and root proteome variations in tomato. Journal of Experimental Botany 62: 2797–2813. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

