Integrated metagenomics/metaproteomics reveals human host-microbiota signatures of Crohn's disease
- PMID: 23209564
- PMCID: PMC3509130
- DOI: 10.1371/journal.pone.0049138
Integrated metagenomics/metaproteomics reveals human host-microbiota signatures of Crohn's disease
Abstract
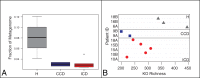
Crohn's disease (CD) is an inflammatory bowel disease of complex etiology, although dysbiosis of the gut microbiota has been implicated in chronic immune-mediated inflammation associated with CD. Here we combined shotgun metagenomic and metaproteomic approaches to identify potential functional signatures of CD in stool samples from six twin pairs that were either healthy, or that had CD in the ileum (ICD) or colon (CCD). Integration of these omics approaches revealed several genes, proteins, and pathways that primarily differentiated ICD from healthy subjects, including depletion of many proteins in ICD. In addition, the ICD phenotype was associated with alterations in bacterial carbohydrate metabolism, bacterial-host interactions, as well as human host-secreted enzymes. This eco-systems biology approach underscores the link between the gut microbiota and functional alterations in the pathophysiology of Crohn's disease and aids in identification of novel diagnostic targets and disease specific biomarkers.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

