Nuclear RNA sequencing of the mouse erythroid cell transcriptome
- PMID: 23209567
- PMCID: PMC3510205
- DOI: 10.1371/journal.pone.0049274
Nuclear RNA sequencing of the mouse erythroid cell transcriptome
Abstract
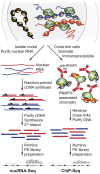
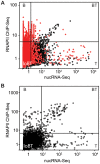
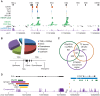
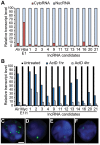
In addition to protein coding genes a substantial proportion of mammalian genomes are transcribed. However, most transcriptome studies investigate steady-state mRNA levels, ignoring a considerable fraction of the transcribed genome. In addition, steady-state mRNA levels are influenced by both transcriptional and posttranscriptional mechanisms, and thus do not provide a clear picture of transcriptional output. Here, using deep sequencing of nuclear RNAs (nucRNA-Seq) in parallel with chromatin immunoprecipitation sequencing (ChIP-Seq) of active RNA polymerase II, we compared the nuclear transcriptome of mouse anemic spleen erythroid cells with polymerase occupancy on a genome-wide scale. We demonstrate that unspliced transcripts quantified by nucRNA-seq correlate with primary transcript frequencies measured by RNA FISH, but differ from steady-state mRNA levels measured by poly(A)-enriched RNA-seq. Highly expressed protein coding genes showed good correlation between RNAPII occupancy and transcriptional output; however, genome-wide we observed a poor correlation between transcriptional output and RNAPII association. This poor correlation is due to intergenic regions associated with RNAPII which correspond with transcription factor bound regulatory regions and a group of stable, nuclear-retained long non-coding transcripts. In conclusion, sequencing the nuclear transcriptome provides an opportunity to investigate the transcriptional landscape in a given cell type through quantification of unspliced primary transcripts and the identification of nuclear-retained long non-coding RNAs.
Conflict of interest statement
Figures



References
-
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, et al. (2005) The transcriptional landscape of the mammalian genome. Science 309: 1559–1563. - PubMed
-
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, et al. (2005) Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308: 1149–1154. - PubMed
-
- Kim JH, Tuziak T, Hu L, Wang Z, Bondaruk J, et al. (2005) Alterations in transcription clusters underlie development of bladder cancer along papillary and nonpapillary pathways. Lab Invest 85: 532–549. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

