Population differentiation of southern Indian male lineages correlates with agricultural expansions predating the caste system
- PMID: 23209694
- PMCID: PMC3508930
- DOI: 10.1371/journal.pone.0050269
Population differentiation of southern Indian male lineages correlates with agricultural expansions predating the caste system
Erratum in
- PLoS One. 2013;8(7). doi:10.1371/annotation/8663819b-5ff0-4133-b70a-2d686dfb0a44
Abstract
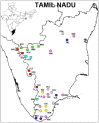
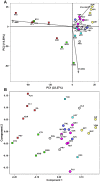
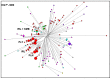
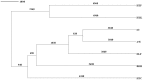
Previous studies that pooled Indian populations from a wide variety of geographical locations, have obtained contradictory conclusions about the processes of the establishment of the Varna caste system and its genetic impact on the origins and demographic histories of Indian populations. To further investigate these questions we took advantage that both Y chromosome and caste designation are paternally inherited, and genotyped 1,680 Y chromosomes representing 12 tribal and 19 non-tribal (caste) endogamous populations from the predominantly Dravidian-speaking Tamil Nadu state in the southernmost part of India. Tribes and castes were both characterized by an overwhelming proportion of putatively Indian autochthonous Y-chromosomal haplogroups (H-M69, F-M89, R1a1-M17, L1-M27, R2-M124, and C5-M356; 81% combined) with a shared genetic heritage dating back to the late Pleistocene (10-30 Kya), suggesting that more recent Holocene migrations from western Eurasia contributed <20% of the male lineages. We found strong evidence for genetic structure, associated primarily with the current mode of subsistence. Coalescence analysis suggested that the social stratification was established 4-6 Kya and there was little admixture during the last 3 Kya, implying a minimal genetic impact of the Varna (caste) system from the historically-documented Brahmin migrations into the area. In contrast, the overall Y-chromosomal patterns, the time depth of population diversifications and the period of differentiation were best explained by the emergence of agricultural technology in South Asia. These results highlight the utility of detailed local genetic studies within India, without prior assumptions about the importance of Varna rank status for population grouping, to obtain new insights into the relative influences of past demographic events for the population structure of the whole of modern India.
Conflict of interest statement
Figures
References
-
- Cavalli-Sforza LL, Menozzi P, Piazza A (1994) The History and Geography of Human Genes. Princeton, NJ.: Princeton University Press.
-
- Majumder PP (2010) The human genetic history of South Asia. Curr Biol 20: R184–187. - PubMed
-
- Singh KS (2002) People of India: An Introduction, Revised edition: Oxford Uniersity Press.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

