Chicken domestication: an updated perspective based on mitochondrial genomes
- PMID: 23211792
- PMCID: PMC3668654
- DOI: 10.1038/hdy.2012.83
Chicken domestication: an updated perspective based on mitochondrial genomes
Abstract
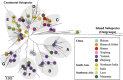
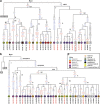
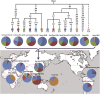
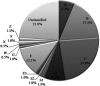
Domestic chickens (Gallus gallus domesticus) fulfill various roles ranging from food and entertainment to religion and ornamentation. To survey its genetic diversity and trace the history of domestication, we investigated a total of 4938 mitochondrial DNA (mtDNA) fragments including 2843 previously published and 2095 de novo units from 2044 domestic chickens and 51 red junglefowl (Gallus gallus). To obtain the highest possible level of molecular resolution, 50 representative samples were further selected for total mtDNA genome sequencing. A fine-gained mtDNA phylogeny was investigated by defining haplogroups A-I and W-Z. Common haplogroups A-G were shared by domestic chickens and red junglefowl. Rare haplogroups H-I and W-Z were specific to domestic chickens and red junglefowl, respectively. We re-evaluated the global mtDNA profiles of chickens. The geographic distribution for each of major haplogroups was examined. Our results revealed new complexities of history in chicken domestication because in the phylogeny lineages from the red junglefowl were mingled with those of the domestic chickens. Several local domestication events in South Asia, Southwest China and Southeast Asia were identified. The assessment of chicken mtDNA data also facilitated our understanding about the Austronesian settlement in the Pacific.
Figures
References
-
- Achilli A, Olivieri A, Pellecchia M, Uboldi C, Colli L, Al-Zahery N, et al. Mitochondrial genomes of extinct aurochs survive in domestic cattle. Curr Biol. 2008;18:R157–R158. - PubMed
-
- Bellwood P.1976Prehistoric plant and animal domestication in AustronesiaIn: GdG Sieveking, Longworth IH, Wilson ED, (eds)Problems in Economic and Social Archaeology Westview Press Boulder, CO; 153–168.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

