MicroRNA 218 acts as a tumor suppressor by targeting multiple cancer phenotype-associated genes in medulloblastoma
- PMID: 23212916
- PMCID: PMC3548500
- DOI: 10.1074/jbc.M112.396762
MicroRNA 218 acts as a tumor suppressor by targeting multiple cancer phenotype-associated genes in medulloblastoma
Abstract
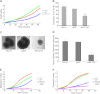
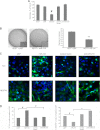
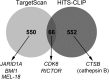
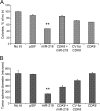
Aberrant expression of microRNAs has been implicated in many cancers. We recently demonstrated differential expression of several microRNAs in medulloblastoma. In this study, the regulation and function of microRNA 218 (miR-218), which is significantly underexpressed in medulloblastoma, was evaluated. Re-expression of miR-218 resulted in a significant decrease in medulloblastoma cell growth, cell colony formation, cell migration, invasion, and tumor sphere size. We used C17.2 neural stem cells as a model to show that increased miR-218 expression results in increased cell differentiation and also decreased malignant transformation when transfected with the oncogene REST. These results suggest that miR-218 acts as a tumor suppressor in medulloblastoma. MicroRNAs function by down-regulating translation of target mRNAs. Targets are determined by imperfect base pairing of the microRNA to the 3'-UTR of the mRNA. To comprehensively identify actual miR-218 targets, medulloblastoma cells overexpressing miR-218 and control cells were subjected to high throughput sequencing of RNA isolated by cross-linking immunoprecipitation, a technique that identifies the mRNAs bound to the RNA-induced silencing complex component protein Argonaute 2. High throughput sequencing of mRNAs identified 618 genes as targets of miR-218 and included both previously validated targets and many targets not predicted computationally. Additional work further confirmed CDK6, RICTOR, and CTSB (cathepsin B) as targets of miR-218 and examined the functional role of one of these targets, CDK6, in medulloblastoma.
Figures


References
-
- Provias J. P., Becker L. E. (1996) Cellular and molecular pathology of medulloblastoma. J. Neurooncol. 29, 35–43 - PubMed
-
- Gilbertson R. J., Ellison D. W. (2008) The origins of medulloblastoma subtypes. Annu. Rev. Pathol. 3, 341–365 - PubMed
-
- Ribi K., Relly C., Landolt M. A., Alber F. D., Boltshauser E., Grotzer M. A. (2005) Outcome of medulloblastoma in children. Long-term complications and quality of life. Neuropediatrics 36, 357–365 - PubMed
-
- Reinhart B. J., Slack F. J., Basson M., Pasquinelli A. E., Bettinger J. C., Rougvie A. E., Horvitz H. R., Ruvkun G. (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906 - PubMed
-
- Pasquinelli A. E., Ruvkun G. (2002) Control of developmental timing by micrornas and their targets. Annu. Rev. Cell Dev. Biol. 18, 495–513 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

