Chemical genetic screen reveals a role for desmosomal adhesion in mammary branching morphogenesis
- PMID: 23212921
- PMCID: PMC3554898
- DOI: 10.1074/jbc.M112.411033
Chemical genetic screen reveals a role for desmosomal adhesion in mammary branching morphogenesis
Abstract
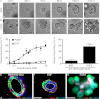
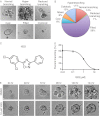
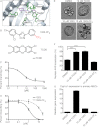
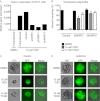
During the process of branching morphogenesis, the mammary gland undergoes distinct phases of remodeling to form an elaborate ductal network that ultimately produces and delivers milk to newborn animals. These developmental events rely on tight regulation of critical cellular pathways, many of which are probably disrupted during initiation and progression of breast cancer. Transgenic mouse and in vitro organoid models previously identified growth factor signaling as a key regulator of mammary branching, but the functional downstream targets of these pathways remain unclear. Here, we used purified primary mammary epithelial cells stimulated with fibroblast growth factor-2 (FGF2) to model mammary branching morphogenesis in vitro. We employed a forward chemical genetic approach to identify modulators of this process and describe a potent compound, 1023, that blocks FGF2-induced branching. In primary mammary epithelial cells, we used lentivirus-mediated knockdown of the aryl hydrocarbon receptor (AHR) to demonstrate that 1023 acts through AHR to block branching. Using 1023 as a tool, we identified desmosomal adhesion as a novel target of AHR signaling and show that desmosomes are critical for AHR agonists to block branching. Our findings support a functional role for desmosomes during mammary morphogenesis and also in blocking FGF-induced invasion.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

