Different secretory repertoires control the biomineralization processes of prism and nacre deposition of the pearl oyster shell
- PMID: 23213212
- PMCID: PMC3529032
- DOI: 10.1073/pnas.1210552109
Different secretory repertoires control the biomineralization processes of prism and nacre deposition of the pearl oyster shell
Abstract
Mollusca evolutionary success can be attributed partly to their efficiency to sustain and protect their soft body with an external biomineralized structure, the shell. Current knowledge of the protein set responsible for the formation of the shell microstructural polymorphism and unique properties remains largely patchy. In Pinctada margaritifera and Pinctada maxima, we identified 80 shell matrix proteins, among which 66 are entirely unique. This is the only description of the whole "biomineralization toolkit" of the matrices that, at least in part, is thought to regulate the formation of the prismatic and nacreous shell layers in the pearl oysters. We unambiguously demonstrate that prisms and nacre are assembled from very different protein repertoires. This suggests that these layers do not derive from each other.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


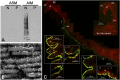
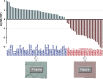

References
-
- Lowenstam AH, Weiner S. On Biomineralization. New York: Oxford Univ Press; 1989.
-
- Killian CE, Wilt FH. Molecular aspects of biomineralization of the echinoderm endoskeleton. Chem Rev. 2008;108(11):4463–4474. - PubMed
-
- Mann S. Molecular recognition in biomineralization. Nature. 1988;332:119–124.
-
- Falini G, Albeck S, Weiner S, Addadi L. Control of aragonite or calcite polymophism by mollusk shell macromolecules. Science. 1996;271:67–69.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

