Reconstitution of the human cytoplasmic dynein complex
- PMID: 23213255
- PMCID: PMC3529016
- DOI: 10.1073/pnas.1210573110
Reconstitution of the human cytoplasmic dynein complex
Abstract
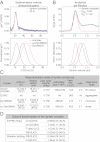
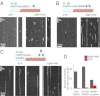
Cytoplasmic dynein is the major motor protein responsible for microtubule minus-end-directed movements in most eukaryotic cells. It transports a variety of cargoes and has numerous functions during spindle assembly and chromosome segregation. It is a large complex of about 1.4 MDa composed of six different subunits, interacting with a multitude of different partners. Most biochemical studies have been performed either with the native mammalian cytoplasmic dynein complex purified from tissue or, more recently, with recombinant dynein fragments from budding yeast and Dictyostelium. Hardly any information exists about the properties of human dynein. Moreover, experiments with an entire human dynein complex prepared from recombinant subunits with a well-defined composition are lacking. Here, we reconstitute a complete cytoplasmic dynein complex using recombinant human subunits and characterize its biochemical and motile properties. Using analytical gel filtration, sedimentation-velocity ultracentrifugation, and negative-stain electron microscopy, we demonstrate that the smaller subunits of the complex have an important structural function for complex integrity. Fluorescence microscopy experiments reveal that while engaged in collective microtubule transport, the recombinant human cytoplasmic dynein complex is an active, microtubule minus-end-directed motor, as expected. However, in contrast to recombinant dynein of nonmetazoans, individual reconstituted human dynein complexes did not show robust processive motility, suggesting a more intricate mechanism of processivity regulation for the human dynein complex. In the future, the comparison of reconstituted dynein complexes from different species promises to provide molecular insight into the mechanisms regulating the various functions of these large molecular machines.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Regulation of processive motion and microtubule localization of cytoplasmic dynein.Biochem Soc Trans. 2015 Feb;43(1):48-57. doi: 10.1042/BST20140252. Biochem Soc Trans. 2015. PMID: 25619245 Review.
-
Three-dimensional structure of cytoplasmic dynein bound to microtubules.Proc Natl Acad Sci U S A. 2007 Dec 26;104(52):20832-7. doi: 10.1073/pnas.0710406105. Epub 2007 Dec 19. Proc Natl Acad Sci U S A. 2007. PMID: 18093913 Free PMC article.
-
DYNLT (Tctex-1) forms a tripartite complex with dynein intermediate chain and RagA, hence linking this small GTPase to the dynein motor.FEBS J. 2015 Oct;282(20):3945-58. doi: 10.1111/febs.13388. Epub 2015 Aug 20. FEBS J. 2015. PMID: 26227614
-
Distinct functions of nucleotide-binding/hydrolysis sites in the four AAA modules of cytoplasmic dynein.Biochemistry. 2004 Sep 7;43(35):11266-74. doi: 10.1021/bi048985a. Biochemistry. 2004. PMID: 15366936
-
Molecular organization and force-generating mechanism of dynein.FEBS J. 2011 Sep;278(17):2964-79. doi: 10.1111/j.1742-4658.2011.08253.x. Epub 2011 Aug 8. FEBS J. 2011. PMID: 21777385 Review.
Cited by
-
Cytoplasmic factories for axonemal dynein assembly.J Cell Sci. 2021 Aug 1;134(15):jcs258626. doi: 10.1242/jcs.258626. Epub 2021 Aug 3. J Cell Sci. 2021. PMID: 34342348 Free PMC article.
-
HEATR5B associates with dynein-dynactin and promotes motility of AP1-bound endosomal membranes.EMBO J. 2023 Dec 1;42(23):e114473. doi: 10.15252/embj.2023114473. Epub 2023 Oct 24. EMBO J. 2023. PMID: 37872872 Free PMC article.
-
Molecular mechanism of dynein-dynactin complex assembly by LIS1.Science. 2024 Mar 29;383(6690):eadk8544. doi: 10.1126/science.adk8544. Epub 2024 Mar 29. Science. 2024. PMID: 38547289 Free PMC article.
-
Two Tetrahymena kinesin-9 family members exhibit slow plus-end-directed motility in vitro.Sci Rep. 2024 Sep 9;14(1):20993. doi: 10.1038/s41598-024-71280-y. Sci Rep. 2024. PMID: 39251704 Free PMC article.
-
Step Sizes and Rate Constants of Single-headed Cytoplasmic Dynein Measured with Optical Tweezers.Sci Rep. 2018 Nov 5;8(1):16333. doi: 10.1038/s41598-018-34549-7. Sci Rep. 2018. PMID: 30397249 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

