The importance of serine 776 in Ataxin-1 partner selection: a FRET analysis
- PMID: 23213356
- PMCID: PMC3513968
- DOI: 10.1038/srep00919
The importance of serine 776 in Ataxin-1 partner selection: a FRET analysis
Abstract
Anomalous expansion of a polymorphic tract in Ataxin-1 causes the autosomal dominant spinocerebellar ataxia type 1. In addition to polyglutamine expansion, requirements for development of pathology are phosphorylation of serine 776 in Ataxin-1 and nuclear localization of the protein. The phosphorylation state of serine 776 is also crucial for selection of the Ataxin-1 multiple partners. Here, we have used FRET for an in cell study of the interaction of Ataxin-1 with the spliceosome-associated U2AF65 and the adaptor 14-3-3 proteins. Using wild-type Ataxin-1 and Ser776 mutants to a phosphomimetic aspartate and to alanine, we show that U2AF65 binds Ataxin-1 in a Ser776 phosphorylation independent manner whereas 14-3-3 interacts with phosphorylated wild-type Ataxin-1 but not with the mutants. These results indicate that Ser776 acts as the molecular switch that discriminates between normal and aberrant function and that phosphomimetics is not a generally valid approach whose applicability should be carefully validated.
Figures

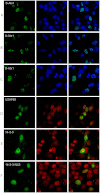
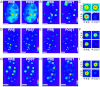



References
-
- Orr H. T., Chung M. Y., Banfi S., Kwiatkowski T. J. Jr., Servadio A., Beaudet A. L., McCall A. E., Duvick L. A., Ranum L. P. & Zoghbi H. Y. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat. Genet. 4, 221–226 (1993). - PubMed
-
- Zoghbi H. Y. & Orr H. T. Spinocerebellar ataxia type 1. Semin. Cell Biol. 6, 29–35 (1995). - PubMed
-
- Matilla-Dueñas A., Goold R. & Giunti P. Clinical, genetic, molecular, and pathophysiological insights into spinocerebellar ataxia type 1. Cerebellum 7, 106–114 (2008). - PubMed
-
- Cummings C. J., Mancini M. A., Antalffy B., DeFranco D. B., Orr H. T. & Zoghbi H. Y. Chaperone suppression of aggregation and altered subcellular proteasome localization imply protein misfolding in SCA1. Nat. Genet. 19, 148–154 (1998). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

