Structural and functional plasticity of subcellular tethering, targeting and processing of RPGRIP1 by RPGR isoforms
- PMID: 23213406
- PMCID: PMC3507198
- DOI: 10.1242/bio.2011489
Structural and functional plasticity of subcellular tethering, targeting and processing of RPGRIP1 by RPGR isoforms
Abstract
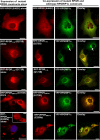
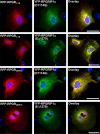
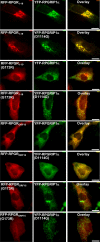
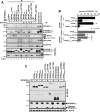
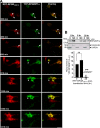
Mutations affecting the retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGRIP1) interactome cause syndromic retinal dystrophies. RPGRIP1 interacts with the retinitis pigmentosa GTPase regulator (RPGR) through a domain homologous to RCC1 (RHD), a nucleotide exchange factor of Ran GTPase. However, functional relationships between RPGR and RPGRIP1 and their subcellular roles are lacking. We show by molecular modeling and analyses of RPGR disease-mutations that the RPGR-interacting domain (RID) of RPGRIP1 embraces multivalently the shared RHD of RPGR(1-19) and RPGR(ORF15) isoforms and the mutations are non-overlapping with the interface found between RCC1 and Ran GTPase. RPGR disease-mutations grouped into six classes based on their structural locations and differential impairment with RPGRIP1 interaction. RPGRIP1α(1) expression alone causes its profuse self-aggregation, an effect suppressed by co-expression of either RPGR isoform before and after RPGRIP1α(1) self-aggregation ensue. RPGR(1-19) localizes to the endoplasmic reticulum, whereas RPGR(ORF15) presents cytosolic distribution and they determine uniquely the subcellular co-localization of RPGRIP1α(1). Disease mutations in RPGR(1) (-19), RPGR(ORF15), or RID of RPGRIP1α(1), singly or in combination, exert distinct effects on the subcellular targeting, co-localization or tethering of RPGRIP1α(1) with RPGR(1-19) or RPGR(ORF15) in kidney, photoreceptor and hepatocyte cell lines. Additionally, RPGR(ORF15), but not RPGR(1-19), protects the RID of RPGRIP1α(1) from limited proteolysis. These studies define RPGR- and cell-type-dependent targeting pathways with structural and functional plasticity modulating the expression of mutations in RPGR and RPGRIP1. Further, RPGR isoforms distinctively determine the subcellular targeting of RPGRIP1α(1,) with deficits in RPGR(ORF15)-dependent intracellular localization of RPGRIP1α(1) contributing to pathomechanisms shared by etiologically distinct syndromic retinal dystrophies.
Keywords: RPGR; RPGRIP1; degeneration; kidney cells; photoreceptor; protein aggregation; protein targeting.
Conflict of interest statement
Figures





References
-
- Abagyan R., Totrov M., Kuznetsov D. (1994). ICM-a new method for protein modeling and design: applications to docking and structure prediction from the distorted native conformation. J. Comput. Chem. 15, 488–506 10.1002/jcc.540150503 - DOI
-
- Abagyan R., Orry A., Raush E., Totrov M. (2010). ICM User Guide 3.7. La Jolla, CA: Molsoft LLC
-
- al-Ubaidi M. R., Font R. L., Quiambao A. B., Keener M. J., Liou G. I., Overbeek P. A., Baehr W. (1992). Bilateral retinal and brain tumors in transgenic mice expressing simian virus 40 large T antigen under control of the human interphotoreceptor retinoid-binding protein promoter. J. Cell Biol. 119, 1681–1687 10.1083/jcb.119.6.1681 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

