Drosophila Syncrip binds the gurken mRNA localisation signal and regulates localised transcripts during axis specification
- PMID: 23213441
- PMCID: PMC3507208
- DOI: 10.1242/bio.2012885
Drosophila Syncrip binds the gurken mRNA localisation signal and regulates localised transcripts during axis specification
Abstract
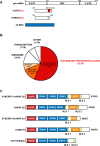
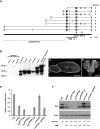
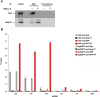
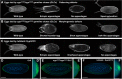
In the Drosophila oocyte, mRNA transport and localised translation play a fundamental role in axis determination and germline formation of the future embryo. gurken mRNA encodes a secreted TGF-α signal that specifies dorsal structures, and is localised to the dorso-anterior corner of the oocyte via a cis-acting 64 nucleotide gurken localisation signal. Using GRNA chromatography, we characterised the biochemical composition of the ribonucleoprotein complexes that form around the gurken mRNA localisation signal in the oocyte. We identified a number of the factors already known to be involved in gurken localisation and translational regulation, such as Squid and Imp, in addition to a number of factors with known links to mRNA localisation, such as Me31B and Exu. We also identified previously uncharacterised Drosophila proteins, including the fly homologue of mammalian SYNCRIP/hnRNPQ, a component of RNA transport granules in the dendrites of mammalian hippocampal neurons. We show that Drosophila Syncrip binds specifically to gurken and oskar, but not bicoid transcripts. The loss-of-function and overexpression phenotypes of syncrip in Drosophila egg chambers show that the protein is required for correct grk and osk mRNA localisation and translational regulation. We conclude that Drosophila Syncrip is a new factor required for localisation and translational regulation of oskar and gurken mRNA in the oocyte. We propose that Syncrip/SYNCRIP is part of a conserved complex associated with localised transcripts and required for their correct translational regulation in flies and mammals.
Keywords: Drosophila; Syncrip; localised translation; mRNA localization; oogenesis.
Conflict of interest statement
Figures

References
-
- Bannai H., Fukatsu K., Mizutani A., Natsume T., Iemura S., Ikegami T., Inoue T., Mikoshiba K. (2004). An RNA-interacting protein, SYNCRIP (heterogeneous nuclear ribonuclear protein Q1/NSAP1) is a component of mRNA granule transported with inositol 1,4,5-trisphosphate receptor type 1 mRNA in neuronal dendrites. J. Biol. Chem. 279, 53427–53434 10.1074/jbc.M409732200 - DOI - PubMed
-
- Barbee S. A., Estes P. S., Cziko A. M., Hillebrand J., Luedeman R. A., Coller J. M., Johnson N., Howlett I. C., Geng C., Ueda R. et al. (2006). Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron 52, 997–1009 10.1016/j.neuron.2006.10.028 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

