MRE11 facilitates the removal of human topoisomerase II complexes from genomic DNA
- PMID: 23213480
- PMCID: PMC3507232
- DOI: 10.1242/bio.20121834
MRE11 facilitates the removal of human topoisomerase II complexes from genomic DNA
Abstract
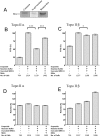
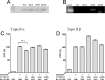
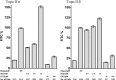
Topoisomerase II creates a double-strand break intermediate with topoisomerase covalently coupled to the DNA via a 5'-phosphotyrosyl bond. These intermediate complexes can become cytotoxic protein-DNA adducts and DSB repair at these lesions requires removal of topoisomerase II. To analyse removal of topoisomerase II from genomic DNA we adapted the trapped in agarose DNA immunostaining assay. Recombinant MRE11 from 2 sources removed topoisomerase IIα from genomic DNA in vitro, as did MRE11 immunoprecipitates isolated from A-TLD or K562 cells. Basal topoisomerase II complex levels were very high in A-TLD cells lacking full-length wild type MRE11, suggesting that MRE11 facilitates the processing of topoisomerase complexes that arise as part of normal cellular metabolism. In K562 cells inhibition of MRE11, PARP or replication increased topoisomerase IIα and β complex levels formed in the absence of an anti-topoisomerase II drug.
Keywords: A-TLD; DSB repair; MRE11; Protein-DNA adducts; Topoisomerase II.
Conflict of interest statement
Figures




References
-
- Alchanati I., Teicher C., Cohen G., Shemesh V., Barr H. M., Nakache P., Ben-Avraham D., Idelevich A., Angel I., Livnah N. et al. (2009). The E3 ubiquitin-ligase Bmi1/Ring1A controls the proteasomal degradation of Top2α cleavage complex - a potentially new drug target. PLoS ONE 4, e8104 10.1371/journal.pone.0008104 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

