Effects of select histidine to cysteine mutations on transcriptional regulation by Escherichia coli RcnR
- PMID: 23215580
- PMCID: PMC3610428
- DOI: 10.1021/bi300886q
Effects of select histidine to cysteine mutations on transcriptional regulation by Escherichia coli RcnR
Abstract
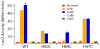
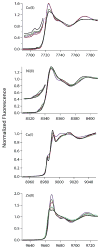
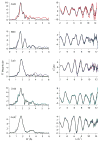
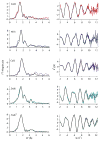
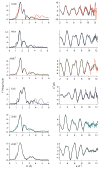
The RcnR metalloregulator represses the transcription of the Co(II) and Ni(II) exporter, RcnAB. Previous studies have shown that Co(II) and Ni(II) bind to RcnR in six-coordinate sites, resulting in derepression. Here, the roles of His60, His64, and His67 in specific metal recognition are examined. His60 and His64 correspond to ligands that are important for Cu(I) binding in the homologous Cu(I)-responsive metalloregulator, CsoR. These residues are known to be functionally important in RcnR transcriptional regulation. X-ray absorption spectroscopy (XAS) was used to examine the structure of bound cognate and noncognate metal ions, and lacZ reporter assays were used to assess the transcription of rcnA in response to metal binding in the three His → Cys mutations, H60C, H64C, and H67C. These studies confirm that both Ni(II) and Co(II) use His64 as a ligand. H64C-RcnR is also the only known mutant that retains a Co(II) response while eliminating the response to Ni(II) binding. XAS data indicate that His60 and His67 are potential Co(II) ligands. The effects of the mutations of His60, His64, and His67 on the structures of the noncognate metal ions [Zn(II) and Cu(I)] reveal that these residues have distinctive roles in binding noncognate metals. None of the His → Cys mutants in RcnR confer any response to Cu(I) binding, including H64C-RcnR, where the ligands involved in Cu(I) binding in CsoR are present. These data indicate that while the secondary, tertiary, and quaternary structures of CsoR and RcnR are quite similar, small changes in primary sequence reveal that the specific mechanisms involved in metal recognition are quite different.
Figures

References
-
- Rosenzweig AC. Metallochaperones: bind and deliver. Chem Biol. 2002;9:673–677. - PubMed
-
- Pennella MA, Giedroc DP. Structural determinants of metal selectivity in prokaryotic metal-responsive transcriptional regulators. BioMetals. 2005;18:413–428. - PubMed
-
- Böck A, King PW, Blokesch M, Posewitz MC. Maturation of hydrogenases. Adv Microb Physiol. 2006;51:1–71. - PubMed
-
- Forzi L, Sawers RG. Maturation of [NiFe]-hydrogenases in Escherichia coli. BioMetals. 2007;20:565–578. - PubMed
-
- De Pina K, Navarro C, McWalter L, Boxer DH, Price NC, Kelly SM, Mandrand-Berthelot MA, Wu LF. Purification and characterization of the periplasmic nickel-binding protein NikA of Escherichia coli K12. Eur J Biochem. 1995;227:857–865. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

