CD25 preselective anti-HIV vectors for improved HIV gene therapy
- PMID: 23216020
- PMCID: PMC4015076
- DOI: 10.1089/hgtb.2012.142
CD25 preselective anti-HIV vectors for improved HIV gene therapy
Abstract
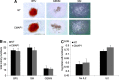
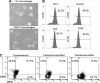
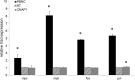
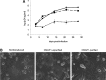
As HIV continues to be a global public health problem with no effective vaccine available, new and innovative therapies, including HIV gene therapies, need to be developed. Due to low transduction efficiencies that lead to low in vivo gene marking, therapeutically relevant efficacy of HIV gene therapy has been difficult to achieve in a clinical setting. Methods to improve the transplantation of enriched populations of anti-HIV vector-transduced cells may greatly increase the in vivo efficacy of HIV gene therapies. Here we describe the development of preselective anti-HIV lentiviral vectors that allow for the purification of vector-transduced cells to achieve an enriched population of HIV-resistant cells. A selectable protein, human CD25, not normally found on CD34+ hematopoietic progenitor cells (HPCs), was incorporated into a triple combination anti-HIV lentiviral vector. Upon purification of cells transduced with the preselective anti-HIV vector, safety was demonstrated in CD34+ HPCs and in HPC-derived macrophages in vitro. Upon challenge with HIV-1, improved efficacy was observed in purified preselective anti-HIV vector-transduced macrophages compared to unpurified cells. These proof-of-concept results highlight the potential use of this method to improve HIV stem cell gene therapy for future clinical applications.
Figures


References
-
- Baldanti F. Paolucci S. Gulminetti R., et al. Early emergence of raltegravir resistance mutations in patients receiving HAART salvage regimens. J. Med. Virol. 2010;82:116–122. - PubMed
-
- Bauer G. Valdez P. Kearns K., et al. Inhibition of human immunodeficiency virus-1 (HIV-1) replication after transduction of granulocyte colony-stimulating factor-mobilized CD34+ cells from HIV-1-infected donors using retroviral vectors containing anti-HIV-1 genes. Blood. 1997;89:2259–2267. - PubMed
-
- Clark F.J. Gregg R. Piper K., et al. Chronic graft-versus-host-disease is associated with increased numbers of peripheral blood CD4+CD25high regulatory T cells. Blood. 2004;103:2410–2416. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

