Biomolecular electrostatics and solvation: a computational perspective
- PMID: 23217364
- PMCID: PMC3533255
- DOI: 10.1017/S003358351200011X
Biomolecular electrostatics and solvation: a computational perspective
Abstract
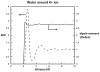
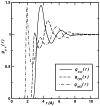
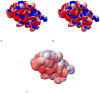
An understanding of molecular interactions is essential for insight into biological systems at the molecular scale. Among the various components of molecular interactions, electrostatics are of special importance because of their long-range nature and their influence on polar or charged molecules, including water, aqueous ions, proteins, nucleic acids, carbohydrates, and membrane lipids. In particular, robust models of electrostatic interactions are essential for understanding the solvation properties of biomolecules and the effects of solvation upon biomolecular folding, binding, enzyme catalysis, and dynamics. Electrostatics, therefore, are of central importance to understanding biomolecular structure and modeling interactions within and among biological molecules. This review discusses the solvation of biomolecules with a computational biophysics view toward describing the phenomenon. While our main focus lies on the computational aspect of the models, we provide an overview of the basic elements of biomolecular solvation (e.g. solvent structure, polarization, ion binding, and non-polar behavior) in order to provide a background to understand the different types of solvation models.
Figures

References
-
- AAQVIST J. Ion-water interaction potentials derived from free energy perturbation simulations. The Journal of Physical Chemistry. 1990;94(21):8021–8024.
-
- ADAMS PL, STAHLEY MR, KOSEK AB, WANG JM, STROBEL SA. Crystal structure of a self-splicing group I intron with both exons. Nature. 2004;430(6995):45–50. - PubMed
-
- ALEJANDRE J, HANSEN J. Ions in water: From ion clustering to crystal nucleation. Physical Review E. 2007;76(6):061505. - PubMed
-
- ALLEN TW, KUYUCAK S, CHUNG SH. Molecular dynamics estimates of ion diffusion in model hydrophobic and KcsA potassium channels. Biophysical Chemistry. 2000;86(1):1–14. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

