The emerging diversity of transpososome architectures
- PMID: 23217365
- PMCID: PMC7292550
- DOI: 10.1017/S0033583512000145
The emerging diversity of transpososome architectures
Abstract
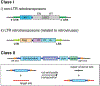
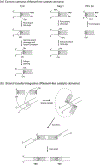
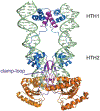
DNA transposases are enzymes that catalyze the movement of discrete pieces of DNA from one location in the genome to another. Transposition occurs through a series of controlled DNA strand cleavage and subsequent integration reactions that are carried out by nucleoprotein complexes known as transpososomes. Transpososomes are dynamic assemblies which must undergo conformational changes that control DNA breaks and ensure that, once started, the transposition reaction goes to completion. They provide a precise architecture within which the chemical reactions involved in transposon movement occur, but adopt different conformational states as transposition progresses. Their components also vary as they must, at some stage, include target DNA and sometimes even host-encoded proteins. A very limited number of transpososome states have been crystallographically captured, and here we provide an overview of the various structures determined to date. These structures include examples of DNA transposases that catalyze transposition by a cut-and-paste mechanism using an RNaseH-like nuclease catalytic domain, those that transpose using only single-stranded DNA substrates and targets, and the retroviral integrases that carry out an integration reaction very similar to DNA transposition. Given that there are a number of common functional requirements for transposition, it is remarkable how these are satisfied by complex assemblies that are so architecturally different.
Figures




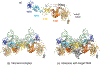

References
-
- Arciszewska LK, Drake D & Craig NL (1989). Transposon Tn7. cis-acting sequences in transposition and transposition immunity. Journal of Molecular Biology 207, 35–52. - PubMed
-
- Arciszewska LK, Mckown RL & Craig NL (1991). Purification of TnsB, a transposition protein that binds to the ends of Tn7. Journal of Biological Chemistry 266, 21736–21744. - PubMed
-
- Augé-Gouillou C, Hamelin MH, Demattei MV, Periquet M & Bigot Y (2001). The wild-type conformation of the Mos-1 inverted terminal repeats is suboptimal for transposition in bacteria. Molecular Genetics and Genomics 265, 51–57. - PubMed
-
- Augé-Gouillou C, Brillet B, Germon S, Hamelin MH & Bigot Y (2005a). Mariner Mos1 transposase dimerizes prior to ITR binding. Journal of Molecular Biology 351, 117–130. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

