Noninvasive imaging beyond the diffraction limit of 3D dynamics in thickly fluorescent specimens
- PMID: 23217717
- PMCID: PMC3615549
- DOI: 10.1016/j.cell.2012.10.008
Noninvasive imaging beyond the diffraction limit of 3D dynamics in thickly fluorescent specimens
Abstract
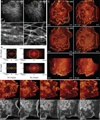
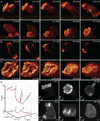
Optical imaging of the dynamics of living specimens involves tradeoffs between spatial resolution, temporal resolution, and phototoxicity, made more difficult in three dimensions. Here, however, we report that rapid three-dimensional (3D) dynamics can be studied beyond the diffraction limit in thick or densely fluorescent living specimens over many time points by combining ultrathin planar illumination produced by scanned Bessel beams with super-resolution structured illumination microscopy. We demonstrate in vivo karyotyping of chromosomes during mitosis and identify different dynamics for the actin cytoskeleton at the dorsal and ventral surfaces of fibroblasts. Compared to spinning disk confocal microscopy, we demonstrate substantially reduced photodamage when imaging rapid morphological changes in D. discoideum cells, as well as improved contrast and resolution at depth within developing C. elegans embryos. Bessel beam structured plane illumination thus promises new insights into complex biological phenomena that require 4D subcellular spatiotemporal detail in either a single or multicellular context.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures





References
-
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. - PubMed
-
- Booth MJ. Adaptive optics in microscopy. Phil. Trans. R. Soc. Lond. Ser. A. 2007;365:2829–2843. - PubMed
-
- Cella Zanachhi F, Lavagnino Z, Perrone Donnorso M, Del Bue A, Furia L, Faretta M, Diaspro A. Live-cell 3D super-resolution imaging in thick biological samples. Nat. Meth. 2011;8:1047–1049. - PubMed
-
- Dupuy D, Bertin N, Hidalgo CA, Venkatesan K, Tu D, Lee D, Rosenberg J, Svrzikapa N, Blanc A, Carnec A, Carvunis A-R, Pulak R, Shingles J, Reece-Hoyes J, Hunt-Newbury R, Viveiros R, Mohler WA, Tasan M, Roth FP, Le Peuch C, Hope IA, Johnsen R, Moerman DG, Barabasi A-L, Baillie D, Vidal M. Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans. Nat. Biotech. 2007;25:663–668. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

