Reversible RNA adenosine methylation in biological regulation
- PMID: 23218460
- PMCID: PMC3558665
- DOI: 10.1016/j.tig.2012.11.003
Reversible RNA adenosine methylation in biological regulation
Abstract
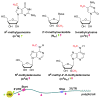
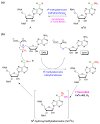
N(6)-methyladenosine (m(6)A) is a ubiquitous modification in mRNA and other RNAs across most eukaryotes. For many years, however, the exact functions of m(6)A were not clearly understood. The discovery that the fat mass and obesity-associated protein (FTO) is an m(6)A demethylase indicates that this modification is reversible and dynamically regulated, suggesting that it has regulatory roles. In addition, it has been shown that m(6)A affects cell fate decisions in yeast and plant development. Recent affinity-based m(6)A profiling in mouse and human cells further showed that this modification is a widespread mark in coding and noncoding RNA (ncRNA) transcripts and is likely dynamically regulated throughout developmental processes. Therefore, reversible RNA methylation, analogous to reversible DNA and histone modifications, may affect gene expression and cell fate decisions by modulating multiple RNA-related cellular pathways, which potentially provides rapid responses to various cellular and environmental signals, including energy and nutrient availability in mammals.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures


References
-
- Sharp PA. The centrality of RNA. Cell. 2009;136:577–580. - PubMed
-
- He C. Grand challenge commentary: RNA epigenetics? Nat Chem Biol. 2010;6:863–865. - PubMed
-
- Rottman FM, et al. Sequences containing methylated nucleotides at the 5′-termini of messenger RNAs: possible implications for processing. Cell. 1974;3:197–199. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

