Translational control at the synapse: role of RNA regulators
- PMID: 23218750
- PMCID: PMC3530003
- DOI: 10.1016/j.tibs.2012.11.001
Translational control at the synapse: role of RNA regulators
Abstract
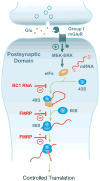
Translational control of gene expression is instrumental in the regulation of eukaryotic cellular form and function. Neurons in particular rely on this form of control because their numerous synaptic connections need to be independently modulated in an input-specific manner. Brain cytoplasmic (BC) RNAs implement translational control at neuronal synapses. BC RNAs regulate protein synthesis by interacting with eIF4 translation initiation factors. Recent evidence suggests that such regulation is required to control synaptic strength, and that dysregulation of local protein synthesis precipitates neuronal hyperexcitability and a propensity for epileptogenic responses. A similar phenotype results from lack of fragile X mental retardation protein (FMRP), indicating that BC RNAs and FMRP use overlapping and convergent modes of action in neuronal translational regulation.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures
Similar articles
-
Fragile X mental retardation protein induces synapse loss through acute postsynaptic translational regulation.J Neurosci. 2007 Mar 21;27(12):3120-30. doi: 10.1523/JNEUROSCI.0054-07.2007. J Neurosci. 2007. PMID: 17376973 Free PMC article.
-
Metabotropic glutamate receptors and fragile x mental retardation protein: partners in translational regulation at the synapse.Sci Signal. 2008 Feb 5;1(5):pe6. doi: 10.1126/stke.15pe6. Sci Signal. 2008. PMID: 18272470 Review.
-
Post-translational modifications of the Fragile X Mental Retardation Protein in neuronal function and dysfunction.Mol Psychiatry. 2020 Aug;25(8):1688-1703. doi: 10.1038/s41380-019-0629-4. Epub 2019 Dec 10. Mol Psychiatry. 2020. PMID: 31822816 Review.
-
Tracking the Fragile X Mental Retardation Protein in a Highly Ordered Neuronal RiboNucleoParticles Population: A Link between Stalled Polyribosomes and RNA Granules.PLoS Genet. 2016 Jul 27;12(7):e1006192. doi: 10.1371/journal.pgen.1006192. eCollection 2016 Jul. PLoS Genet. 2016. PMID: 27462983 Free PMC article.
-
Impaired activity-dependent FMRP translation and enhanced mGluR-dependent LTD in Fragile X premutation mice.Hum Mol Genet. 2013 Mar 15;22(6):1180-92. doi: 10.1093/hmg/dds525. Epub 2012 Dec 18. Hum Mol Genet. 2013. PMID: 23250915 Free PMC article.
Cited by
-
Long-term memory consolidation: The role of RNA-binding proteins with prion-like domains.RNA Biol. 2017 May 4;14(5):568-586. doi: 10.1080/15476286.2016.1244588. Epub 2016 Oct 11. RNA Biol. 2017. PMID: 27726526 Free PMC article. Review.
-
ncRNA BC1 influences translation in the oocyte.RNA Biol. 2021 Nov;18(11):1893-1904. doi: 10.1080/15476286.2021.1880181. Epub 2021 Feb 8. RNA Biol. 2021. PMID: 33491548 Free PMC article.
-
Regulatory BC200 RNA in peripheral blood of patients with invasive breast cancer.J Investig Med. 2018 Oct;66(7):1055-1063. doi: 10.1136/jim-2018-000717. Epub 2018 Jul 2. J Investig Med. 2018. PMID: 29967012 Free PMC article.
-
Subcellular fractionation and localization studies reveal a direct interaction of the fragile X mental retardation protein (FMRP) with nucleolin.PLoS One. 2014 Mar 21;9(3):e91465. doi: 10.1371/journal.pone.0091465. eCollection 2014. PLoS One. 2014. PMID: 24658146 Free PMC article.
-
Knockdown of BC200 RNA expression reduces cell migration and invasion by destabilizing mRNA for calcium-binding protein S100A11.RNA Biol. 2017 Oct 3;14(10):1418-1430. doi: 10.1080/15476286.2017.1297913. Epub 2017 Mar 1. RNA Biol. 2017. PMID: 28277927 Free PMC article.
References
-
- Mathews MB, et al., editors. Translational control in biology and medicine. Cold Spring Harbor Laboratory Press; 2007.
-
- Job C, Eberwine J. Localization and translation of mRNA in dendrites and axons. Nat Rev Neurosci. 2001;2:889–898. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

