Potent inhibition of feline coronaviruses with peptidyl compounds targeting coronavirus 3C-like protease
- PMID: 23219425
- PMCID: PMC3563934
- DOI: 10.1016/j.antiviral.2012.11.005
Potent inhibition of feline coronaviruses with peptidyl compounds targeting coronavirus 3C-like protease
Abstract
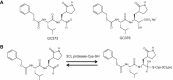
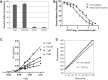
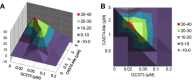
Feline coronavirus infection is common among domestic and exotic felid species and usually associated with mild or asymptomatic enteritis; however, feline infectious peritonitis (FIP) is a fatal disease of cats that is caused by systemic infection with a feline infectious peritonitis virus (FIPV), a variant of feline enteric coronavirus (FECV). Currently, there is no specific treatment approved for FIP despite the importance of FIP as the leading infectious cause of death in young cats. During the replication process, coronavirus produces viral polyproteins that are processed into mature proteins by viral proteases, the main protease (3C-like [3CL] protease) and the papain-like protease. Since the cleavages of viral polyproteins are an essential step for virus replication, blockage of viral protease is an attractive target for therapeutic intervention. Previously, we reported the generation of broad-spectrum peptidyl inhibitors against viruses that possess a 3C or 3CL protease. In this study, we further evaluated the antiviral effects of the peptidyl inhibitors against feline coronaviruses, and investigated the interaction between our protease inhibitor and a cathepsin B inhibitor, an entry blocker, against a feline coronavirus in cell culture. Herein we report that our compounds behave as reversible, competitive inhibitors of 3CL protease, potently inhibited the replication of feline coronaviruses (EC(50) in a nanomolar range) and, furthermore, combination of cathepsin B and 3CL protease inhibitors led to a strong synergistic interaction against feline coronaviruses in a cell culture system.
Copyright © 2012 Elsevier B.V. All rights reserved.
Figures


References
-
- Barlough J.E., Shacklett B.L. Antiviral studies of feline infectious peritonitis virus in vitro. Vet. Rec. 1994;135:177–179. - PubMed
-
- Chang H.W., de Groot R.J., Egberink H.F., Rottier J.M. Feline infectious peritonitis: insights into feline coronavirus pathobiogenesis and epidemiology based on genetic analysis of the viral 3c gene. J. Gen. Virology. 2010;91:415–420. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

