Biotinylation of lysine 16 in histone H4 contributes toward nucleosome condensation
- PMID: 23219734
- PMCID: PMC3563257
- DOI: 10.1016/j.abb.2012.11.005
Biotinylation of lysine 16 in histone H4 contributes toward nucleosome condensation
Abstract
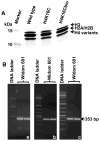
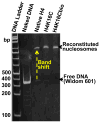
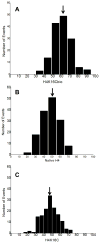
Holocarboxylase synthetase (HLCS) is part of a multiprotein gene repression complex and catalyzes the covalent binding of biotin to lysines (K) in histones H3 and H4, thereby creating rare gene repression marks such as K16-biotinylated histone H4 (H4K16bio). We tested the hypothesis that H4K16bio contributes toward nucleosome condensation and gene repression by HLCS. We used recombinant histone H4 in which K16 was mutated to a cysteine (H4K16C) for subsequent chemical biotinylation of the sulfhydryl group to create H4K16Cbio. Nucleosomes were assembled by using H4K16Cbio and the 'Widom 601' nucleosomal DNA position sequence; biotin-free histone H4 and H4K16C were used as controls. Nucleosomal compaction was analyzed using atomic force microscopy (AFM). The length of DNA per nucleosome was ∼30% greater in H4K16Cbio-containing histone octamers (61.14±10.92nm) compared with native H4 (46.89±12.6nm) and H4K16C (47.26±10.32nm), suggesting biotin-dependent chromatin condensation (P<0.001). Likewise, the number of DNA turns around histone core octamers was ∼17.2% greater in in H4K16Cbio-containing octamers (1.78±0.16) compared with native H4 (1.52±0.21) and H4K16C (1.52±0.17), judged by the rotation angle (P<0.001; N=150). We conclude that biotinylation of K16 in histone H4 contributes toward chromatin condensation.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures


References
-
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–60. - PubMed
-
- Andrews AJ, Luger K. A coupled equilibrium approach to study nucleosome thermodynamics. Methods Enzymol. 2011;488:265–85. - PubMed
-
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. - PubMed
-
- Stanley JS, Griffin JB, Zempleni J. Biotinylation of histones in human cells: effects of cell proliferation. Eur J Biochem. 2001;268:5424–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

