Three-dimensional mRNA measurements reveal minimal regional heterogeneity in esophageal squamous cell carcinoma
- PMID: 23219752
- PMCID: PMC3562732
- DOI: 10.1016/j.ajpath.2012.10.028
Three-dimensional mRNA measurements reveal minimal regional heterogeneity in esophageal squamous cell carcinoma
Abstract
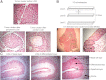
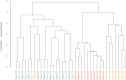
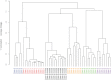
The classic tumor clonal evolution theory postulates that cancers change over time to produce unique molecular subclones within a parent neoplasm, presumably including regional differences in gene expression. More recently, however, this notion has been challenged by studies showing that tumors maintain a relatively stable transcript profile. To examine these competing hypotheses, we microdissected discrete subregions containing approximately 3000 to 8000 cells (500 to 1500 μm in diameter) from ex vivo esophageal squamous cell carcinoma (ESCC) specimens and analyzed transcriptomes throughout three-dimensional tumor space. Overall mRNA profiles were highly similar in all 59 intratumor comparisons, in distinct contrast to the markedly different global expression patterns observed in other dissected cell populations. For example, normal esophageal basal cells contained 1918 and 624 differentially expressed genes at a greater than twofold level (95% confidence level of <5% false positives), compared with normal differentiated esophageal cells and ESCC, respectively. In contrast, intratumor regions had only zero to four gene changes at a greater than twofold level, with most tumor comparisons showing none. The present data indicate that, when analyzed using a standard array-based method at this level of histological resolution, ESCC contains little regional mRNA heterogeneity.
Copyright © 2013 American Society for Investigative Pathology. Published by Elsevier Inc. All rights reserved.
Figures
References
-
- Kamangar F., Dores G.M., Anderson W.F. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24:2137–2150. - PubMed
-
- Parkin D.M., Bray F., Ferlay J., Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. - PubMed
-
- Jemal A., Siegel R., Xu J., Ward E. Cancer Statistics, 2010. CA Cancer J Clin. 2010;60:277–300. [Erratum appeared in CA Cancer J Clin 2011, 61:133–134] - PubMed
-
- Pisani P., Bray F., Parkin D.M. Estimates of the world-wide prevalence of cancer for 25 sites in the adult population. Int J Cancer. 2002;97:72–81. - PubMed
-
- Travis W.D. Pathology of lung cancer. Clin Chest Med. 2002;23:65–81. viii. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

