The eFIP system for text mining of protein interaction networks of phosphorylated proteins
- PMID: 23221174
- PMCID: PMC3514748
- DOI: 10.1093/database/bas044
The eFIP system for text mining of protein interaction networks of phosphorylated proteins
Abstract
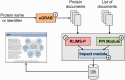
Protein phosphorylation is a central regulatory mechanism in signal transduction involved in most biological processes. Phosphorylation of a protein may lead to activation or repression of its activity, alternative subcellular location and interaction with different binding partners. Extracting this type of information from scientific literature is critical for connecting phosphorylated proteins with kinases and interaction partners, along with their functional outcomes, for knowledge discovery from phosphorylation protein networks. We have developed the Extracting Functional Impact of Phosphorylation (eFIP) text mining system, which combines several natural language processing techniques to find relevant abstracts mentioning phosphorylation of a given protein together with indications of protein-protein interactions (PPIs) and potential evidences for impact of phosphorylation on the PPIs. eFIP integrates our previously developed tools, Extracting Gene Related ABstracts (eGRAB) for document retrieval and name disambiguation, Rule-based LIterature Mining System (RLIMS-P) for Protein Phosphorylation for extraction of phosphorylation information, a PPI module to detect PPIs involving phosphorylated proteins and an impact module for relation extraction. The text mining system has been integrated into the curation workflow of the Protein Ontology (PRO) to capture knowledge about phosphorylated proteins. The eFIP web interface accepts gene/protein names or identifiers, or PubMed identifiers as input, and displays results as a ranked list of abstracts with sentence evidence and summary table, which can be exported in a spreadsheet upon result validation. As a participant in the BioCreative-2012 Interactive Text Mining track, the performance of eFIP was evaluated on document retrieval (F-measures of 78-100%), sentence-level information extraction (F-measures of 70-80%) and document ranking (normalized discounted cumulative gain measures of 93-100% and mean average precision of 0.86). The utility and usability of the eFIP web interface were also evaluated during the BioCreative Workshop. The use of the eFIP interface provided a significant speed-up (∼2.5-fold) for time to completion of the curation task. Additionally, eFIP significantly simplifies the task of finding relevant articles on PPI involving phosphorylated forms of a given protein.
Figures


References
-
- Chen L, Liu H, Friedman C. Gene name ambiguity of eukaryotic nomenclatures. Bioinformatics. 2005;21:248–256. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

