Hypervariable domains of nsP3 proteins of New World and Old World alphaviruses mediate formation of distinct, virus-specific protein complexes
- PMID: 23221551
- PMCID: PMC3571466
- DOI: 10.1128/JVI.02853-12
Hypervariable domains of nsP3 proteins of New World and Old World alphaviruses mediate formation of distinct, virus-specific protein complexes
Abstract
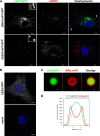
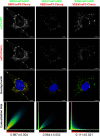
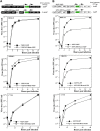
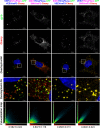
Alphaviruses are a group of single-stranded RNA viruses with genomes of positive polarity. They are divided into two geographically isolated groups: the Old World and the New World alphaviruses. Despite their similar genome organizations and virion structures, they differ in many aspects of pathogenesis and interaction with the host cell. Here we present new data highlighting previously unknown differences between these two groups. We found that nsP3 proteins of Sindbis virus (SINV) and Venezuelan equine encephalitis virus (VEEV) form cytoplasmic complexes with different morphologies and protein compositions. Unlike the amorphous aggregates formed by SINV nsP3 and other Old World alphavirus-specific nsP3s, VEEV nsP3 forms unique, large spherical structures with striking symmetry. Moreover, VEEV nsP3 does not interact with proteins previously identified as major components of SINV nsP3 complexes, such as G3BP1 and G3BP2. Importantly, the morphology of the complexes and the specificity of the interaction with cellular proteins are largely determined by the hypervariable domain (HVD) of nsP3. Replacement of the VEEV nsP3 HVD with the corresponding domain of SINV nsP3 rendered this protein capable of interaction with G3BPs. Conversely, replacement of the SINV nsP3 HVD with that of VEEV abolished SINV nsP3's interaction with G3BPs. The replacement of natural HVDs with those from heterologous viruses did not abrogate virus replication, despite these fragments demonstrating very low levels of sequence identity. Our data suggest that in spite of the differences in morphology and composition of the SINV- and VEEV-specific nsP3 complexes, it is likely that they have similar functions in virus replication and modification of the cellular environment.
Figures



References
-
- Rulli NE, Melton J, Wilmes A, Ewart G, Mahalingam S. 2007. The molecular and cellular aspects of arthritis due to alphavirus infections: lesson learned from Ross River virus. Ann. N. Y. Acad. Sci. 1102:96–108 - PubMed
-
- Staples JE, Breiman RF, Powers AM. 2009. Chikungunya fever: an epidemiological review of a re-emerging infectious disease. Clin. Infect. Dis. 49:942–948 - PubMed
-
- Toivanen A. 2008. Alphaviruses: an emerging cause of arthritis? Curr. Opin. Rheumatol. 20:486–490 - PubMed
-
- Weaver SC, Ferro C, Barrera R, Boshell J, Navarro JC. 2004. Venezuelan equine encephalitis. Annu. Rev. Entomol. 49:141–174 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

