Design requirements for interfering particles to maintain coadaptive stability with HIV-1
- PMID: 23221552
- PMCID: PMC3571494
- DOI: 10.1128/JVI.02741-12
Design requirements for interfering particles to maintain coadaptive stability with HIV-1
Abstract
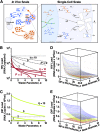
Defective interfering particles (DIPs) are viral deletion mutants lacking essential transacting or packaging elements and must be complemented by wild-type virus to propagate. DIPs transmit through human populations, replicating at the expense of the wild-type virus and acting as molecular parasites of viruses. Consequently, engineered DIPs have been proposed as therapies for a number of diseases, including human immunodeficiency virus (HIV). However, it is not clear if DIP-based therapies would face evolutionary blocks given the high mutation rates and high within-host diversity of lentiviruses. Divergent evolution of HIV and DIPs appears likely since natural DIPs have not been detected for lentiviruses, despite extensive sequencing of HIVs and simian immunodeficiency viruses (SIVs). Here, we tested if the apparent lack of lentiviral DIPs is due to natural selection and analyzed which molecular characteristics a DIP or DIP-based therapy would need to maintain coadaptive stability with HIV-1. Using a well-established mathematical model of HIV-1 in a host extended to include its replication in a single cell and interference from DIP, we calculated evolutionary selection coefficients. The analysis predicts that interference by codimerization between DIPs and HIV-1 genomes is evolutionarily unstable, indicating that recombination between DIPs and HIV-1 would be selected against. In contrast, DIPs that interfere via competition for capsids have the potential to be evolutionarily stable if the capsid-to-genome production ratio of HIV-1 is >1. Thus, HIV-1 variants that attempt to "starve" DIPs to escape interference would be selected against. In summary, the analysis suggests specific experimental measurements that could address the apparent lack of naturally occurring lentiviral DIPs and specifies how therapeutic approaches based on engineered DIPs could be evolutionarily robust and avoid recombination.
Figures

Comment in
-
Coadaptive stability of interfering particles with HIV-1 when there is an evolutionary conflict.J Virol. 2013 Sep;87(17):9959. doi: 10.1128/JVI.00705-13. J Virol. 2013. PMID: 23943739 Free PMC article. No abstract available.
-
Reply to "Coadaptive stability of interfering particles with HIV-1 when there is an evolutionary conflict".J Virol. 2013 Sep;87(17):9960-2. doi: 10.1128/JVI.00932-13. J Virol. 2013. PMID: 23943740 Free PMC article. No abstract available.
Similar articles
-
High-Throughput Single-Cell Kinetics of Virus Infections in the Presence of Defective Interfering Particles.J Virol. 2015 Nov 25;90(3):1599-612. doi: 10.1128/JVI.02190-15. Print 2016 Feb 1. J Virol. 2015. PMID: 26608322 Free PMC article.
-
Modeling the intracellular replication of influenza A virus in the presence of defective interfering RNAs.Virus Res. 2016 Feb 2;213:90-99. doi: 10.1016/j.virusres.2015.11.016. Epub 2015 Nov 23. Virus Res. 2016. PMID: 26592173
-
Coadaptive stability of interfering particles with HIV-1 when there is an evolutionary conflict.J Virol. 2013 Sep;87(17):9959. doi: 10.1128/JVI.00705-13. J Virol. 2013. PMID: 23943739 Free PMC article. No abstract available.
-
Antivirotics based on defective interfering particles: emerging concepts and challenges.Front Cell Infect Microbiol. 2025 Feb 24;15:1436026. doi: 10.3389/fcimb.2025.1436026. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40066067 Free PMC article. Review.
-
The defective component of viral populations.Curr Opin Virol. 2018 Dec;33:74-80. doi: 10.1016/j.coviro.2018.07.014. Epub 2018 Aug 9. Curr Opin Virol. 2018. PMID: 30099321 Review.
Cited by
-
Reply to "Coadaptive stability of interfering particles with HIV-1 when there is an evolutionary conflict".J Virol. 2013 Sep;87(17):9960-2. doi: 10.1128/JVI.00932-13. J Virol. 2013. PMID: 23943740 Free PMC article. No abstract available.
-
Single-Cell Analysis Uncovers a Vast Diversity in Intracellular Viral Defective Interfering RNA Content Affecting the Large Cell-to-Cell Heterogeneity in Influenza A Virus Replication.Viruses. 2020 Jan 7;12(1):71. doi: 10.3390/v12010071. Viruses. 2020. PMID: 31936115 Free PMC article.
-
Continuous influenza virus production in cell culture shows a periodic accumulation of defective interfering particles.PLoS One. 2013 Sep 5;8(9):e72288. doi: 10.1371/journal.pone.0072288. eCollection 2013. PLoS One. 2013. PMID: 24039749 Free PMC article.
-
Antigenic evolution of viruses in host populations.PLoS Pathog. 2018 Sep 12;14(9):e1007291. doi: 10.1371/journal.ppat.1007291. eCollection 2018 Sep. PLoS Pathog. 2018. PMID: 30208108 Free PMC article.
-
An evolution-based high-fidelity method of epistasis measurement: Theory and application to influenza.PLoS Pathog. 2021 Jun 21;17(6):e1009669. doi: 10.1371/journal.ppat.1009669. eCollection 2021 Jun. PLoS Pathog. 2021. PMID: 34153082 Free PMC article.
References
-
- Holland J. 1990. Generation and replication of defective viral genomes, p 77–99 In Fields BN, Knipe DM. (ed), Virology. Raven Press, New York, NY
-
- Huang AS, Baltimore D. 1970. Defective viral particles and viral disease processes. Nature 226:325–327 - PubMed
-
- Holland JJ, Villarreal LP. 1975. Purification of defective interfering T particles of vesicular stomatitis and rabies viruses generated in vivo in brains of newborn mice. Virology 67:438–449 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

