A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis
- PMID: 23221598
- PMCID: PMC3556966
- DOI: 10.1105/tpc.112.105163
A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis
Abstract
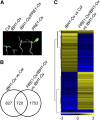
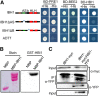
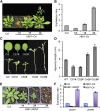
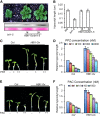
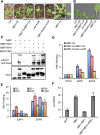
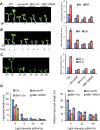
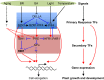
Environmental and endogenous signals, including light, temperature, brassinosteroid (BR), and gibberellin (GA), regulate cell elongation largely by influencing the expression of the paclobutrazol-resistant (PRE) family helix-loop-helix (HLH) factors, which promote cell elongation by interacting antagonistically with another HLH factor, IBH1. However, the molecular mechanism by which PREs and IBH1 regulate gene expression has remained unknown. Here, we show that IBH1 interacts with and inhibits a DNA binding basic helix-loop-helix (bHLH) protein, HBI1, in Arabidopsis thaliana. Overexpression of HBI1 increased hypocotyl and petiole elongation, whereas dominant inactivation of HBI1 and its homologs caused a dwarf phenotype, indicating that HBI1 is a positive regulator of cell elongation. In vitro and in vivo experiments showed that HBI1 directly bound to the promoters and activated two EXPANSIN genes encoding cell wall-loosening enzymes; HBI1's DNA binding and transcriptional activities were inhibited by IBH1, but the inhibitory effects of IBH1 were abolished by PRE1. The results indicate that PREs activate the DNA binding bHLH factor HBI1 by sequestering its inhibitor IBH1. Altering each of the three factors affected plant sensitivities to BR, GA, temperature, and light. Our study demonstrates that PREs, IBH1, and HBI1 form a chain of antagonistic switches that regulates cell elongation downstream of multiple external and endogenous signals.
Figures

Comment in
-
A tripartite growth regulatory cascade of basic helix-loop-helix transcription factors.Plant Cell. 2012 Dec;24(12):4775. doi: 10.1105/tpc.112.241210. Epub 2012 Dec 7. Plant Cell. 2012. PMID: 23221594 Free PMC article. No abstract available.
References
-
- Carretero-Paulet L., Galstyan A., Roig-Villanova I., Martínez-García J.F., Bilbao-Castro J.R., Robertson D.L. (2010). Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 153: 1398–1412 - PMC - PubMed

