Deficiency of FRAS1-related extracellular matrix 1 (FREM1) causes congenital diaphragmatic hernia in humans and mice
- PMID: 23221805
- PMCID: PMC3561915
- DOI: 10.1093/hmg/dds507
Deficiency of FRAS1-related extracellular matrix 1 (FREM1) causes congenital diaphragmatic hernia in humans and mice
Erratum in
-
Deficiency of FRAS1-related extracellular matrix 1 (FREM1) causes congenital diaphragmatic hernia in humans and mice.Hum Mol Genet. 2020 Apr 15;29(6):1054. doi: 10.1093/hmg/ddz307. Hum Mol Genet. 2020. PMID: 32016392 Free PMC article. No abstract available.
Abstract
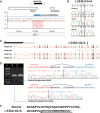
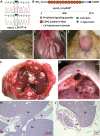
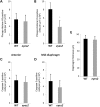
Congenital diaphragmatic hernia (CDH) is a common life-threatening birth defect. Recessive mutations in the FRAS1-related extracellular matrix 1 (FREM1) gene have been shown to cause bifid nose with or without anorectal and renal anomalies (BNAR) syndrome and Manitoba oculotrichoanal (MOTA) syndrome, but have not been previously implicated in the development of CDH. We have identified a female child with an isolated left-sided posterolateral CDH covered by a membranous sac who had no features suggestive of BNAR or MOTA syndromes. This child carries a maternally-inherited ~86 kb FREM1 deletion that affects the expression of FREM1's full-length transcripts and a paternally-inherited splice site mutation that causes activation of a cryptic splice site, leading to a shift in the reading frame and premature termination of all forms of the FREM1 protein. This suggests that recessive FREM1 mutations can cause isolated CDH in humans. Further evidence for the role of FREM1 in the development of CDH comes from an N-ethyl-N-nitrosourea -derived mouse strain, eyes2, which has a homozygous truncating mutation in Frem1. Frem1(eyes2) mice have eye defects, renal agenesis and develop retrosternal diaphragmatic hernias which are covered by a membranous sac. We confirmed that Frem1 is expressed in the anterior portion of the developing diaphragm and found that Frem1(eyes2) embryos had decreased levels of cell proliferation in their developing diaphragms when compared to wild-type embryos. We conclude that FREM1 plays a critical role in the development of the diaphragm and that FREM1 deficiency can cause CDH in both humans and mice.
Figures

Similar articles
-
The role of FREM2 and FRAS1 in the development of congenital diaphragmatic hernia.Hum Mol Genet. 2018 Jun 15;27(12):2064-2075. doi: 10.1093/hmg/ddy110. Hum Mol Genet. 2018. PMID: 29618029 Free PMC article.
-
Gene Expression of FRAS1-Related Extracellular Matrix 1 Is Decreased in Nitrofen-Induced Congenital Diaphragmatic Hernia.Eur J Pediatr Surg. 2016 Feb;26(1):81-5. doi: 10.1055/s-0035-1559884. Epub 2015 Sep 18. Eur J Pediatr Surg. 2016. PMID: 26382659
-
Novel frem1-related mouse phenotypes and evidence of genetic interactions with gata4 and slit3.PLoS One. 2013;8(3):e58830. doi: 10.1371/journal.pone.0058830. Epub 2013 Mar 11. PLoS One. 2013. PMID: 23536828 Free PMC article.
-
Molecular genetics of congenital diaphragmatic defects.Ann Med. 2007;39(4):261-74. doi: 10.1080/07853890701326883. Ann Med. 2007. PMID: 17558598 Free PMC article. Review.
-
Genetic aspects of human congenital diaphragmatic hernia.Clin Genet. 2008 Jul;74(1):1-15. doi: 10.1111/j.1399-0004.2008.01031.x. Epub 2008 May 28. Clin Genet. 2008. PMID: 18510546 Free PMC article. Review.
Cited by
-
The role of FREM2 and FRAS1 in the development of congenital diaphragmatic hernia.Hum Mol Genet. 2018 Jun 15;27(12):2064-2075. doi: 10.1093/hmg/ddy110. Hum Mol Genet. 2018. PMID: 29618029 Free PMC article.
-
Congenital diaphragmatic hernias: from genes to mechanisms to therapies.Dis Model Mech. 2017 Aug 1;10(8):955-970. doi: 10.1242/dmm.028365. Dis Model Mech. 2017. PMID: 28768736 Free PMC article. Review.
-
Unraveling the Genetics of Congenital Diaphragmatic Hernia: An Ongoing Challenge.Front Pediatr. 2022 Feb 3;9:800915. doi: 10.3389/fped.2021.800915. eCollection 2021. Front Pediatr. 2022. PMID: 35186825 Free PMC article. Review.
-
Evidence for an association between Coffin-Siris syndrome and congenital diaphragmatic hernia.Am J Med Genet A. 2022 Sep;188(9):2718-2723. doi: 10.1002/ajmg.a.62889. Epub 2022 Jul 7. Am J Med Genet A. 2022. PMID: 35796094 Free PMC article. Review.
-
Toll-like Interleukin 1 Receptor Regulator Is an Important Modulator of Inflammation Responsive Genes.Front Immunol. 2019 Feb 28;10:272. doi: 10.3389/fimmu.2019.00272. eCollection 2019. Front Immunol. 2019. PMID: 30873160 Free PMC article.
References
-
- Langham M.R., Jr., Kays D.W., Ledbetter D.J., Frentzen B., Sanford L.L., Richards D.S. Congenital diaphragmatic hernia. Epidemiology and outcome. Clin. Perinatol. 1996;23:671–688. - PubMed
-
- Bollmann R., Kalache K., Mau H., Chaoui R., Tennstedt C. Associated malformations and chromosomal defects in congenital diaphragmatic hernia. Fetal Diagn. Ther. 1995;10:52–59. - PubMed
-
- Tibboel D., Gaag A.V. Etiologic and genetic factors in congenital diaphragmatic hernia. Clin. Perinatol. 1996;23:689–699. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

