Naturally selecting solutions: the use of genetic algorithms in bioinformatics
- PMID: 23222169
- PMCID: PMC3813526
- DOI: 10.4161/bioe.23041
Naturally selecting solutions: the use of genetic algorithms in bioinformatics
Abstract
For decades, computer scientists have looked to nature for biologically inspired solutions to computational problems; ranging from robotic control to scheduling optimization. Paradoxically, as we move deeper into the post-genomics era, the reverse is occurring, as biologists and bioinformaticians look to computational techniques, to solve a variety of biological problems. One of the most common biologically inspired techniques are genetic algorithms (GAs), which take the Darwinian concept of natural selection as the driving force behind systems for solving real world problems, including those in the bioinformatics domain. Herein, we provide an overview of genetic algorithms and survey some of the most recent applications of this approach to bioinformatics based problems.
Keywords: genetic algorithm; multiple sequence alignment; optimization; protein structure prediction.
Figures
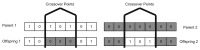

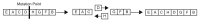
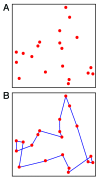
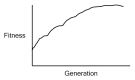




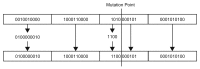
Similar articles
-
Genetic algorithms and their application to in silico evolution of genetic regulatory networks.Methods Mol Biol. 2010;673:297-321. doi: 10.1007/978-1-60761-842-3_19. Methods Mol Biol. 2010. PMID: 20835807
-
The hierarchical fair competition (HFC) framework for sustainable evolutionary algorithms.Evol Comput. 2005 Summer;13(2):241-77. doi: 10.1162/1063656054088530. Evol Comput. 2005. PMID: 15969902
-
A new parallel DNA algorithm to solve the task scheduling problem based on inspired computational model.Biosystems. 2017 Dec;162:59-65. doi: 10.1016/j.biosystems.2017.09.001. Epub 2017 Sep 7. Biosystems. 2017. PMID: 28890344
-
Applications of genetic programming in cancer research.Int J Biochem Cell Biol. 2009 Feb;41(2):405-13. doi: 10.1016/j.biocel.2008.09.025. Epub 2008 Oct 2. Int J Biochem Cell Biol. 2009. PMID: 18929677 Free PMC article. Review.
-
A review on multiple sequence alignment from the perspective of genetic algorithm.Genomics. 2017 Oct;109(5-6):419-431. doi: 10.1016/j.ygeno.2017.06.007. Epub 2017 Jun 29. Genomics. 2017. PMID: 28669847 Review.
Cited by
-
A beginner's guide to phylogenetics.Microb Ecol. 2013 Jul;66(1):1-4. doi: 10.1007/s00248-013-0236-x. Epub 2013 Apr 28. Microb Ecol. 2013. PMID: 23624570 Review.
-
A transcriptomic study for identifying cardia- and non-cardia-specific gastric cancer prognostic factors using genetic algorithm-based methods.J Cell Mol Med. 2020 Aug;24(16):9457-9465. doi: 10.1111/jcmm.15618. Epub 2020 Jul 10. J Cell Mol Med. 2020. PMID: 32649057 Free PMC article.
-
Membrane protein orientation and refinement using a knowledge-based statistical potential.BMC Bioinformatics. 2013 Sep 18;14:276. doi: 10.1186/1471-2105-14-276. BMC Bioinformatics. 2013. PMID: 24047460 Free PMC article.
-
The superior fault tolerance of artificial neural network training with a fault/noise injection-based genetic algorithm.Protein Cell. 2016 Oct;7(10):735-748. doi: 10.1007/s13238-016-0302-5. Epub 2016 Aug 9. Protein Cell. 2016. PMID: 27502185 Free PMC article.
-
GA-GBLUP: leveraging the genetic algorithm to improve the predictability of genomic selection.Brief Bioinform. 2024 Jul 25;25(5):bbae385. doi: 10.1093/bib/bbae385. Brief Bioinform. 2024. PMID: 39101500 Free PMC article.
References
-
- Holland JH. Adaptation in natural and artiðcial systems. Ann Arbor: University of Michigan Press 1975; 2.
-
- Kumar SN, Panneerselvam R. A Survey on the Vehicle Routing Problem and Its Variants. Intelligent Information Management. 2012;4:66–74. doi: 10.4236/iim.2012.43010. - DOI
-
- Gemperline P, Niazi A, Leardi R. Genetic algorithms in chemometrics. J Chemometr. 2012;26:345–51. doi: 10.1002/cem.2426. - DOI
-
- Sharma RN, Pancholi SS. Optimization techniques in pharmaceutical industry: A Review. Journal of Current Pharmaceutical Research. 2011;7:21–8.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
