TET2 promotes histone O-GlcNAcylation during gene transcription
- PMID: 23222540
- PMCID: PMC3684361
- DOI: 10.1038/nature11742
TET2 promotes histone O-GlcNAcylation during gene transcription
Abstract
Ten eleven translocation (TET) enzymes, including TET1, TET2 and TET3, convert 5-methylcytosine to 5-hydroxymethylcytosine and regulate gene transcription. However, the molecular mechanism by which TET family enzymes regulate gene transcription remains elusive. Using protein affinity purification, here we search for functional partners of TET proteins, and find that TET2 and TET3 associate with O-linked β-N-acetylglucosamine (O-GlcNAc) transferase (OGT), an enzyme that by itself catalyses the addition of O-GlcNAc onto serine and threonine residues (O-GlcNAcylation) in vivo. TET2 directly interacts with OGT, which is important for the chromatin association of OGT in vivo. Although this specific interaction does not regulate the enzymatic activity of TET2, it facilitates OGT-dependent histone O-GlcNAcylation. Moreover, OGT associates with TET2 at transcription start sites. Downregulation of TET2 reduces the amount of histone 2B Ser 112 GlcNAc marks in vivo, which are associated with gene transcription regulation. Taken together, these results reveal a TET2-dependent O-GlcNAcylation of chromatin. The double epigenetic modifications on both DNA and histones by TET2 and OGT coordinate together for the regulation of gene transcription.
Figures

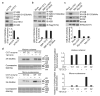
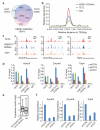
Comment in
-
Chromatin: TET2 keeps histones sweet.Nat Rev Mol Cell Biol. 2013 Feb;14(2):64-5. doi: 10.1038/nrm3511. Epub 2013 Jan 17. Nat Rev Mol Cell Biol. 2013. PMID: 23325359 No abstract available.
-
O-GlcNAcylation and 5-methylcytosine oxidation: an unexpected association between OGT and TETs.Mol Cell. 2013 Feb 21;49(4):618-9. doi: 10.1016/j.molcel.2013.02.006. Mol Cell. 2013. PMID: 23438858 Free PMC article.
References
-
- Ficz G, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. - PubMed
References for full methods
-
- Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nat Protoc. 2007;2:1445–1457. - PubMed
-
- Fujiki R, et al. GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis. Nature. 2009;459:455–459. - PubMed
-
- Yang Z, et al. MicroRNA hsa-miR-138 inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells through adenovirus EID-1. Stem Cells Dev. 2011;20:259–267. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

