FMNL3 FH2-actin structure gives insight into formin-mediated actin nucleation and elongation
- PMID: 23222643
- PMCID: PMC3876896
- DOI: 10.1038/nsmb.2462
FMNL3 FH2-actin structure gives insight into formin-mediated actin nucleation and elongation
Abstract
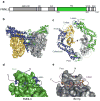
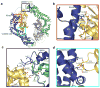
Formins are actin-assembly factors that act in a variety of actin-based processes. The conserved formin homology 2 (FH2) domain promotes filament nucleation and influences elongation through interaction with the barbed end. FMNL3 is a formin that induces assembly of filopodia but whose FH2 domain is a poor nucleator. The 3.4-Å structure of a mouse FMNL3 FH2 dimer in complex with tetramethylrhodamine-actin uncovers details of formin-regulated actin elongation. We observe distinct FH2 actin-binding regions; interactions in the knob and coiled-coil subdomains are necessary for actin binding, whereas those in the lasso-post interface are important for the stepping mechanism. Biochemical and cellular experiments test the importance of individual residues for function. This structure provides details for FH2-mediated filament elongation by processive capping and supports a model in which C-terminal non-FH2 residues of FMNL3 are required to stabilize the filament nucleus.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
-
- Goode BL, Eck MJ. Mechanism and function of formins in the control of actin assembly. Annual review of biochemistry. 2007;76:593–627. - PubMed
-
- Xu Y, et al. Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell. 2004;116:711–23. - PubMed
-
- Otomo T, et al. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433:488–94. - PubMed
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

