Differential analysis of gene regulation at transcript resolution with RNA-seq
- PMID: 23222703
- PMCID: PMC3869392
- DOI: 10.1038/nbt.2450
Differential analysis of gene regulation at transcript resolution with RNA-seq
Abstract
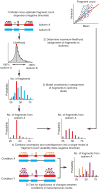
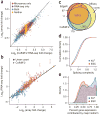
Differential analysis of gene and transcript expression using high-throughput RNA sequencing (RNA-seq) is complicated by several sources of measurement variability and poses numerous statistical challenges. We present Cuffdiff 2, an algorithm that estimates expression at transcript-level resolution and controls for variability evident across replicate libraries. Cuffdiff 2 robustly identifies differentially expressed transcripts and genes and reveals differential splicing and promoter-preference changes. We demonstrate the accuracy of our approach through differential analysis of lung fibroblasts in response to loss of the developmental transcription factor HOXA1, which we show is required for lung fibroblast and HeLa cell cycle progression. Loss of HOXA1 results in significant expression level changes in thousands of individual transcripts, along with isoform switching events in key regulators of the cell cycle. Cuffdiff 2 performs robust differential analysis in RNA-seq experiments at transcript resolution, revealing a layer of regulation not readily observable with other high-throughput technologies.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

