Ferric-pyoverdine recognition by Fpv outer membrane proteins of Pseudomonas protegens Pf-5
- PMID: 23222724
- PMCID: PMC3562110
- DOI: 10.1128/JB.01639-12
Ferric-pyoverdine recognition by Fpv outer membrane proteins of Pseudomonas protegens Pf-5
Abstract
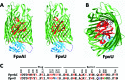
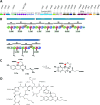
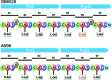
The soil bacterium Pseudomonas protegens Pf-5 (previously called P. fluorescens Pf-5) produces two siderophores, enantio-pyochelin and a compound in the large and diverse pyoverdine family. Using high-resolution mass spectroscopy, we determined the structure of the pyoverdine produced by Pf-5. In addition to producing its own siderophores, Pf-5 also utilizes ferric complexes of some pyoverdines produced by other strains of Pseudomonas spp. as sources of iron. Previously, phylogenetic analysis of the 45 TonB-dependent outer membrane proteins in Pf-5 indicated that six are in a well-supported clade with ferric-pyoverdine receptors (Fpvs) from other Pseudomonas spp. We used a combination of phylogenetics, bioinformatics, mutagenesis, pyoverdine structural determinations, and cross-feeding bioassays to assign specific ferric-pyoverdine substrates to each of the six Fpvs of Pf-5. We identified at least one ferric-pyoverdine that was taken up by each of the six Fpvs of Pf-5. Functional redundancy of the Pf-5 Fpvs was also apparent, with some ferric-pyoverdines taken up by all mutants with a single Fpv deletion but not by a mutant having deletions in two of the Fpv-encoding genes. Finally, we demonstrated that phylogenetically related Fpvs take up ferric complexes of structurally related pyoverdines, thereby establishing structure-function relationships that can be employed in the future to predict the pyoverdine substrates of Fpvs in other Pseudomonas spp.
Figures


Similar articles
-
TonB-dependent outer-membrane proteins and siderophore utilization in Pseudomonas fluorescens Pf-5.Biometals. 2011 Apr;24(2):193-213. doi: 10.1007/s10534-010-9385-2. Epub 2010 Nov 16. Biometals. 2011. PMID: 21080032
-
Genomic, genetic and structural analysis of pyoverdine-mediated iron acquisition in the plant growth-promoting bacterium Pseudomonas fluorescens SBW25.BMC Microbiol. 2008 Jan 14;8:7. doi: 10.1186/1471-2180-8-7. BMC Microbiol. 2008. PMID: 18194565 Free PMC article.
-
Siderophore-mediated iron uptake in fluorescent Pseudomonas: characterization of the pyoverdine-receptor binding site of three cross-reacting pyoverdines.Arch Biochem Biophys. 2002 Jan 15;397(2):179-83. doi: 10.1006/abbi.2001.2667. Arch Biochem Biophys. 2002. PMID: 11795869
-
Pyoverdines: pigments, siderophores and potential taxonomic markers of fluorescent Pseudomonas species.Arch Microbiol. 2000 Sep;174(3):135-42. doi: 10.1007/s002030000188. Arch Microbiol. 2000. PMID: 11041343 Review.
-
Diversity of siderophore-mediated iron uptake systems in fluorescent pseudomonads: not only pyoverdines.Environ Microbiol. 2002 Dec;4(12):787-98. doi: 10.1046/j.1462-2920.2002.00369.x. Environ Microbiol. 2002. PMID: 12534462 Review.
Cited by
-
The genome of a steinernematid-associated Pseudomonas piscis bacterium encodes the biosynthesis of insect toxins.Access Microbiol. 2023 Oct 5;5(10):000659.v3. doi: 10.1099/acmi.0.000659.v3. eCollection 2023. Access Microbiol. 2023. PMID: 37970093 Free PMC article.
-
PvdN Enzyme Catalyzes a Periplasmic Pyoverdine Modification.J Biol Chem. 2016 Nov 11;291(46):23929-23938. doi: 10.1074/jbc.M116.755611. Epub 2016 Oct 4. J Biol Chem. 2016. PMID: 27703013 Free PMC article.
-
Effect of tannic acid on the transcriptome of the soil bacterium Pseudomonas protegens Pf-5.Appl Environ Microbiol. 2013 May;79(9):3141-5. doi: 10.1128/AEM.03101-12. Epub 2013 Feb 22. Appl Environ Microbiol. 2013. PMID: 23435890 Free PMC article.
-
The biosynthesis of pyoverdines.Microb Cell. 2018 Aug 28;5(10):424-437. doi: 10.15698/mic2018.10.649. Microb Cell. 2018. PMID: 30386787 Free PMC article. Review.
-
Total substitution and partial modification of the set of non-ribosomal peptide synthetases clusters lead to pyoverdine diversity in the Pseudomonas fluorescens complex.Front Microbiol. 2024 Aug 19;15:1421749. doi: 10.3389/fmicb.2024.1421749. eCollection 2024. Front Microbiol. 2024. PMID: 39224222 Free PMC article.
References
-
- Ramette A, Frapolli M, Fischer-La Saux M, Gruffaz C, Meyer JM, Défago G, Sutra L, Moënne-Loccoz Y. 2011. Pseudomonas protegens sp. nov., widespread plant-protecting bacteria producing the biocontrol compounds 2,4-diacetylphloroglucinol and pyoluteorin. Syst. Appl. Microbiol. 34: 180–188 - PubMed
-
- Howell CR, Stipanovic RD. 1979. Control of Rhizoctonia solani in cotton seedlings with Pseudomonas fluorescens and with an antibiotic produced by the bacterium. Phytopathology 69: 480–482
-
- Loper JE, Paulsen IT, Kobayashi DY. 2007. The genomic sequence of Pseudomonas fluorescens Pf-5: insights into biological control. Phytopathology 97: 233–238 - PubMed
-
- Youard ZA, Mislin GL, Majcherczyk PA, Schalk IJ, Reimmann C. 2007. Pseudomonas fluorescens CHA0 produces enantio-pyochelin, the optical antipode of the Pseudomonas aeruginosa siderophore pyochelin. J. Biol. Chem. 282: 35546–35553 - PubMed
-
- Budzikiewicz H. 2004. Siderophores of the Pseudomonadaceae sensu stricto (fluorescent and non-fluorescent Pseudomonas spp.). Fortschr. Chem. Org. Naturst. 87: 81–237 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

