Chlamydia trachomatis outer membrane complex protein B (OmcB) is processed by the protease CPAF
- PMID: 23222729
- PMCID: PMC3571324
- DOI: 10.1128/JB.02087-12
Chlamydia trachomatis outer membrane complex protein B (OmcB) is processed by the protease CPAF
Abstract
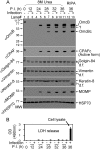
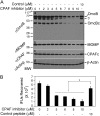
We previously reported that the Chlamydia trachomatis outer membrane complex protein B (OmcB) was partially processed in Chlamydia-infected cells. We have now confirmed that the OmcB processing occurred inside live cells during chlamydial infection and was not due to proteolysis during sample harvesting. OmcB processing was preceded by the generation of active CPAF, a serine protease known to be able to cross the inner membrane via a Sec-dependent pathway, suggesting that active CPAF is available for processing OmcB in the periplasm. In a cell-free system, CPAF activity is both necessary and sufficient for processing OmcB. Both depletion of CPAF from Chlamydia-infected cell lysates with a CPAF-specific antibody and blocking CPAF activity with a CPAF-specific inhibitory peptide removed the OmcB processing ability of the lysates. A highly purified wild-type CPAF but not a catalytic residue-substituted mutant CPAF was sufficient for processing OmcB. Most importantly, in chlamydial culture, inhibition of CPAF with a specific inhibitory peptide blocked OmcB processing and reduced the recovery of infectious organisms. Thus, we have identified OmcB as a novel authentic target for the putative chlamydial virulence factor CPAF, which should facilitate our understanding of the roles of CPAF in chlamydial biology and pathogenesis.
Figures


Similar articles
-
A Chlamydia trachomatis OmcB C-terminal fragment is released into the host cell cytoplasm and is immunogenic in humans.Infect Immun. 2011 Jun;79(6):2193-203. doi: 10.1128/IAI.00003-11. Epub 2011 Mar 21. Infect Immun. 2011. PMID: 21422182 Free PMC article.
-
CPAF: a Chlamydial protease in search of an authentic substrate.PLoS Pathog. 2012;8(8):e1002842. doi: 10.1371/journal.ppat.1002842. Epub 2012 Aug 2. PLoS Pathog. 2012. PMID: 22876181 Free PMC article.
-
Characterization of CPAF critical residues and secretion during Chlamydia trachomatis infection.Infect Immun. 2015 Jun;83(6):2234-41. doi: 10.1128/IAI.00275-15. Epub 2015 Mar 16. Infect Immun. 2015. PMID: 25776755 Free PMC article.
-
Chlamydial protease-like activity factor--insights into immunity and vaccine development.J Reprod Immunol. 2009 Dec;83(1-2):179-84. doi: 10.1016/j.jri.2009.05.007. Epub 2009 Oct 23. J Reprod Immunol. 2009. PMID: 19853923 Free PMC article. Review.
-
[Chlamydia trachomatis proteasome protein as one of the significant pathogenicity factors of exciter].Mol Gen Mikrobiol Virusol. 2014;(2):3-8. Mol Gen Mikrobiol Virusol. 2014. PMID: 25080811 Review. Russian.
Cited by
-
Chlamydial Lytic Exit from Host Cells Is Plasmid Regulated.mBio. 2015 Nov 10;6(6):e01648-15. doi: 10.1128/mBio.01648-15. mBio. 2015. PMID: 26556273 Free PMC article.
-
3-O Sulfated Heparan Sulfate (G2) Peptide Ligand Impairs the Infectivity of Chlamydia muridarum.Biomolecules. 2025 Jul 12;15(7):999. doi: 10.3390/biom15070999. Biomolecules. 2025. PMID: 40723871 Free PMC article.
-
TargeTron Inactivation of Chlamydia trachomatis gseA Results in a Lipopolysaccharide 3-Deoxy-d-Manno-Oct-2-Ulosonic Acid-Deficient Strain That Is Cytotoxic for Cells.Infect Immun. 2023 Jul 18;91(7):e0009623. doi: 10.1128/iai.00096-23. Epub 2023 May 31. Infect Immun. 2023. PMID: 37255490 Free PMC article.
-
Reassessing the role of the secreted protease CPAF in Chlamydia trachomatis infection through genetic approaches.Pathog Dis. 2014 Aug;71(3):336-51. doi: 10.1111/2049-632X.12179. Epub 2014 May 16. Pathog Dis. 2014. PMID: 24838663 Free PMC article.
-
The multiple functions of the numerous Chlamydia trachomatis secreted proteins: the tip of the iceberg.Microb Cell. 2019 Aug 21;6(9):414-449. doi: 10.15698/mic2019.09.691. Microb Cell. 2019. PMID: 31528632 Free PMC article. Review.
References
-
- Centers for Disease Control and Prevention 2009. Sexually transmitted disease surveillance, 2008 Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/std/stats08/toc.htm
-
- Land JA, Van Bergen JE, Morre SA, Postma MJ. 2010. Epidemiology of Chlamydia trachomatis infection in women and the cost-effectiveness of screening. Hum. Reprod. Update 16:189–204 - PubMed
-
- Stephens RS. 2003. The cellular paradigm of chlamydial pathogenesis. Trends Microbiol. 11:44–51 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

