Somatic neurofibromatosis type 1 (NF1) inactivation characterizes NF1-associated pilocytic astrocytoma
- PMID: 23222849
- PMCID: PMC3589532
- DOI: 10.1101/gr.142604.112
Somatic neurofibromatosis type 1 (NF1) inactivation characterizes NF1-associated pilocytic astrocytoma
Abstract
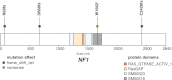
Low-grade brain tumors (pilocytic astrocytomas) arising in the neurofibromatosis type 1 (NF1) inherited cancer predisposition syndrome are hypothesized to result from a combination of germline and acquired somatic NF1 tumor suppressor gene mutations. However, genetically engineered mice (GEM) in which mono-allelic germline Nf1 gene loss is coupled with bi-allelic somatic (glial progenitor cell) Nf1 gene inactivation develop brain tumors that do not fully recapitulate the neuropathological features of the human condition. These observations raise the intriguing possibility that, while loss of neurofibromin function is necessary for NF1-associated low-grade astrocytoma development, additional genetic changes may be required for full penetrance of the human brain tumor phenotype. To identify these potential cooperating genetic mutations, we performed whole-genome sequencing (WGS) analysis of three NF1-associated pilocytic astrocytoma (PA) tumors. We found that the mechanism of somatic NF1 loss was different in each tumor (frameshift mutation, loss of heterozygosity, and methylation). In addition, tumor purity analysis revealed that these tumors had a high proportion of stromal cells, such that only 50%-60% of cells in the tumor mass exhibited somatic NF1 loss. Importantly, we identified no additional recurrent pathogenic somatic mutations, supporting a model in which neuroglial progenitor cell NF1 loss is likely sufficient for PA formation in cooperation with a proper stromal environment.
Figures


References
-
- Bajenaru ML, Hernandez MR, Perry A, Zhu Y, Parada LF, Garbow JR, Gutmann DH 2003. Optic nerve glioma in mice requires astrocyte Nf1 gene inactivation and Nf1 brain heterozygosity. Cancer Res 63: 8573–8577 - PubMed
-
- Bajenaru ML, Garbow JR, Perry A, Hernandez MR, Gutmann DH 2005. Natural history of neurofibromatosis 1-associated optic nerve glioma in mice. Ann Neurol 57: 119–127 - PubMed
-
- Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, Delano D, Zhang L, Schroth GP, Gunderson KL, et al. 2011. High density DNA methylation array with single CpG site resolution. Genomics 98: 288–295 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
