The zinc cluster protein Sut1 contributes to filamentation in Saccharomyces cerevisiae
- PMID: 23223039
- PMCID: PMC3571311
- DOI: 10.1128/EC.00214-12
The zinc cluster protein Sut1 contributes to filamentation in Saccharomyces cerevisiae
Abstract
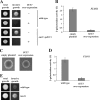
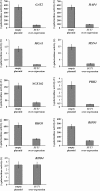
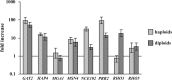
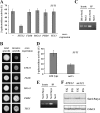
Sut1 is a transcriptional regulator of the Zn(II)(2)Cys(6) family in the budding yeast Saccharomyces cerevisiae. The only function that has been attributed to Sut1 is sterol uptake under anaerobic conditions. Here, we show that Sut1 is also expressed in the presence of oxygen, and we identify a novel function for Sut1. SUT1 overexpression blocks filamentous growth, a response to nutrient limitation, in both haploid and diploid cells. This inhibition by Sut1 is independent of its function in sterol uptake. Sut1 downregulates the expression of GAT2, HAP4, MGA1, MSN4, NCE102, PRR2, RHO3, and RHO5. Several of these Sut1 targets (GAT2, HAP4, MGA1, RHO3, and RHO5) are essential for filamentation in haploids and/or diploids. Furthermore, the expression of the Sut1 target genes, with the exception of MGA1, is induced during filamentous growth. We also show that SUT1 expression is autoregulated and inhibited by Ste12, a key transcriptional regulator of filamentation. We propose that Sut1 partially represses the expression of GAT2, HAP4, MGA1, MSN4, NCE102, PRR2, RHO3, and RHO5 when nutrients are plentiful. Filamentation-inducing conditions relieve this repression by Sut1, and the increased expression of Sut1 targets triggers filamentous growth.
Figures




Similar articles
-
Regulation of mating in the budding yeast Saccharomyces cerevisiae by the zinc cluster proteins Sut1 and Sut2.Biochem Biophys Res Commun. 2013 Aug 16;438(1):66-70. doi: 10.1016/j.bbrc.2013.07.027. Epub 2013 Jul 19. Biochem Biophys Res Commun. 2013. PMID: 23872066
-
The zinc cluster proteins Upc2 and Ecm22 promote filamentation in Saccharomyces cerevisiae by sterol biosynthesis-dependent and -independent pathways.Mol Microbiol. 2016 Feb;99(3):512-27. doi: 10.1111/mmi.13244. Epub 2015 Nov 5. Mol Microbiol. 2016. PMID: 26448198
-
SUT1 is a putative Zn[II]2Cys6-transcription factor whose upregulation enhances both sterol uptake and synthesis in aerobically growing Saccharomyces cerevisiae cells.Eur J Biochem. 2001 Mar;268(6):1585-95. Eur J Biochem. 2001. PMID: 11248676
-
From Lipid Homeostasis to Differentiation: Old and New Functions of the Zinc Cluster Proteins Ecm22, Upc2, Sut1 and Sut2.Int J Mol Sci. 2017 Apr 5;18(4):772. doi: 10.3390/ijms18040772. Int J Mol Sci. 2017. PMID: 28379181 Free PMC article. Review.
-
Copper-regulatory domain involved in gene expression.Prog Nucleic Acid Res Mol Biol. 1998;58:165-95. doi: 10.1016/s0079-6603(08)60036-7. Prog Nucleic Acid Res Mol Biol. 1998. PMID: 9308366 Review.
Cited by
-
Adaptive Response of Saccharomyces Hosts to Totiviridae L-A dsRNA Viruses Is Achieved through Intrinsically Balanced Action of Targeted Transcription Factors.J Fungi (Basel). 2022 Apr 9;8(4):381. doi: 10.3390/jof8040381. J Fungi (Basel). 2022. PMID: 35448612 Free PMC article.
-
Functional divergence of a global regulatory complex governing fungal filamentation.PLoS Genet. 2019 Jan 7;15(1):e1007901. doi: 10.1371/journal.pgen.1007901. eCollection 2019 Jan. PLoS Genet. 2019. PMID: 30615616 Free PMC article.
-
Erg25 Controls Host-Cholesterol Uptake Mediated by Aus1p-Associated Sterol-Rich Membrane Domains in Candida glabrata.Front Cell Dev Biol. 2022 Mar 24;10:820675. doi: 10.3389/fcell.2022.820675. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35399500 Free PMC article.
-
Role of MCC/Eisosome in Fungal Lipid Homeostasis.Biomolecules. 2019 Jul 25;9(8):305. doi: 10.3390/biom9080305. Biomolecules. 2019. PMID: 31349700 Free PMC article. Review.
-
Leveraging transcription factors to speed cellobiose fermentation by Saccharomyces cerevisiae.Biotechnol Biofuels. 2014 Aug 27;7(1):126. doi: 10.1186/s13068-014-0126-6. eCollection 2014. Biotechnol Biofuels. 2014. PMID: 25435910 Free PMC article.
References
-
- Ness F, Bourot S, Régnacq M, Spagnoli R, Bergès T, Karst F. 2001. SUT1 is a putative Zn[II]2Cys6-transcription factor whose upregulation enhances both sterol uptake and synthesis in aerobically growing Saccharomyces cerevisiae cells. Eur. J. Biochem. 268:1585–1595 - PubMed
-
- Bourot S, Karst F. 1995. Isolation and characterization of the Saccharomyces cerevisiae SUT1 gene involved in sterol uptake. Gene 165:97–102 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

