BCR-ABL1 compound mutations in tyrosine kinase inhibitor-resistant CML: frequency and clonal relationships
- PMID: 23223358
- PMCID: PMC3548169
- DOI: 10.1182/blood-2012-05-431379
BCR-ABL1 compound mutations in tyrosine kinase inhibitor-resistant CML: frequency and clonal relationships
Abstract
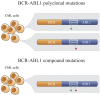
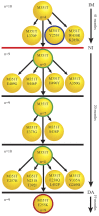
BCR-ABL1 compound mutations can confer high-level resistance to imatinib and other ABL1 tyrosine kinase inhibitors (TKIs). The third-generation ABL1 TKI ponatinib is effective against BCR-ABL1 point mutants individually, but remains vulnerable to certain BCR-ABL1 compound mutants. To determine the frequency of compound mutations among chronic myeloid leukemia patients on ABL1 TKI therapy, in the present study, we examined a collection of patient samples (N = 47) with clear evidence of 2 BCR-ABL1 kinase domain mutations by direct sequencing. Using a cloning and sequencing method, we found that 70% (33/47) of double mutations detected by direct sequencing were compound mutations. Sequential, branching, and parallel routes to compound mutations were common. In addition, our approach revealed individual and compound mutations not detectable by direct sequencing. The frequency of clones harboring compound mutations with more than 2 missense mutations was low (10%), whereas the likelihood of silent mutations increased disproportionately with the total number of mutations per clone, suggesting a limited tolerance for BCR-ABL1 kinase domain missense mutations. We conclude that compound mutations are common in patients with sequencing evidence for 2 BCR-ABL1 mutations and frequently reflect a highly complex clonal network, the evolution of which may be limited by the negative impact of missense mutations on kinase function.
Figures



References
-
- Kantarjian H, Shah NP, Hochhaus A, et al. Dasatinib versus imatinib in newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med. 2010;362(24):2260–2270. - PubMed
-
- Saglio G, Kim DW, Issaragrisil S, et al. Nilotinib versus imatinib for newly diagnosed chronic myeloid leukemia. N Engl J Med. 2010;362(24):2251–2259. - PubMed
-
- Druker B, Guilhot F, O'Brien S, et al. Five-year follow-up of imatinib therapy for newly diagnosed chronic myelogenous leukemia in chronic-phase shows sustained responses and high overall survival. N Engl J Med. 2006;355(23):2408–2417. - PubMed
-
- de Lavallade H, Apperley JF, Khorashad JS, et al. Imatinib for newly diagnosed patients with chronic myeloid leukaemia: incidence of sustained responses in an intention-to-treat analysis. J Clin Oncol. 2008;26(20):3358–3363. - PubMed
-
- Hughes TP, Kaeda J, Branford S, et al. Frequency of major molecular responses to imatinib or interferon alfa plus cytarabine in newly diagnosed chronic myeloid leukemia. N Engl J Med. 2003;349(15):1423–1432. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

