Genotypic prediction of HIV-1 CRF01-AE tropism
- PMID: 23224099
- PMCID: PMC3553889
- DOI: 10.1128/JCM.02328-12
Genotypic prediction of HIV-1 CRF01-AE tropism
Abstract
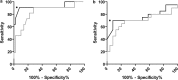
HIV-1 subtype CRF01-AE predominates in south Asia and has spread throughout the world. The virus tropism must be determined before using CCR5 antagonists. Genotypic methods could be used, but the prediction algorithms may be inaccurate for non-B subtypes like CRF01-AE and the correlation with the phenotypic approach has not been assessed. We analyzed 61 CRF01-AE V3 clonal sequences of known phenotype from the GenBank database. The sensitivity of the Geno2pheno10 genotypic algorithm was 91%, but its specificity was poor (54%). In contrast, the combined 11/25 and net charge rule was highly specific (98%) but rather insensitive (64%). We thus identified subtype CRF01-AE determinants in the V3 region that are associated with CXCR4 use and developed a new simple rule for optimizing the genotypic prediction of CRF01-AE tropism. The concordance between the predicted CRF01-AE genotype and the phenotype was 95% for the clonal data set. We then validated this algorithm by analyzing the data from 44 patients infected with subtype CRF01-AE, whose tropism was determined using a recombinant phenotypic entry assay and V3-loop bulk sequencing. The CRF01-AE genotypic tool was 70% sensitive and 96% specific for predicting CXCR4 use, and the concordance between genotype and phenotype was 84%, approaching the concordance obtained for predicting the tropism of HIV-1 subtype B. Genotypic predictions that use a subtype CRF01-AE-specific algorithm appear to be preferable for characterizing coreceptor usage both in pathophysiological studies and for ensuring the appropriate use of CCR5 antagonists.
Figures


References
-
- Berger EA, Murphy PM, Farber JM. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657–700 - PubMed
-
- Dorr P, Westby M, Dobbs S, Griffin P, Irvine B, Macartney M, Mori J, Rickett G, Smith-Burchnell C, Napier C, Webster R, Armour D, Price D, Stammen B, Wood A, Perros M. 2005. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob. Agents Chemother. 49:4721–4732 - PMC - PubMed
-
- Raymond S, Delobel P, Mavigner M, Cazabat M, Souyris C, Sandres-Saune K, Cuzin L, Marchou B, Massip P, Izopet J. 2008. Correlation between genotypic predictions based on V3 sequences and phenotypic determination of HIV-1 tropism. AIDS 22:F11–F16 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

