Relationship of GW/P-bodies with stress granules
- PMID: 23224972
- PMCID: PMC4317337
- DOI: 10.1007/978-1-4614-5107-5_12
Relationship of GW/P-bodies with stress granules
Abstract
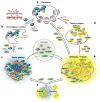
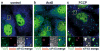
Whereas P-bodies are intimately linked to the cytoplasmic RNA decay machinery, stress granules harbor stalled translation initiation complexes that accumulate upon stress-induced translation arrest. In this Chapter, we reflect on the relationship between P-bodies and stress granules. In mammalian cells, the two structures can be clearly distinguished from each other using specific protein or RNA markers, but they also share many proteins and mRNAs. While the formation of P-bodies and stress granules is coordinately triggered by stress, their assembly appears to be regulated independently by different pathways. Under certain types of stress, P-bodies frequently dock with stress granules, and overexpressing certain proteins that localize to both structures can cause P-body/stress granule fusion. Currently available data suggest that these self-assembling compartments are controlled by flux of mRNAs within the cytoplasm, and that their assembly mirrors the translation and degradation rates of their component mRNAs.
Figures
References
-
- Arimoto K, Fukuda H, Imajoh-Ohmi S, Saito H, Takekawa M. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat Cell Biol. 2008;10:1324–1332. - PubMed
-
- Athanasopoulos V, Barker A, Yu D, Tan AH, Srivastava M, Contreras N, Wang J, Lam KP, Brown SH, Goodnow CC, Dixon NE, Leedman PJ, Saint R, Vinuesa CG. The ROQUIN family of proteins localizes to stress granules via the ROQ domain and binds target mRNAs. FEBS J. 2010;277:2109–2127. - PubMed
-
- Baez MV, Boccaccio GL. Mammalian Smaug is a translational repressor that forms cytoplasmic foci similar to stress granules. J Biol Chem. 2005;280:43131–43140. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

