Halogen bonding (X-bonding): a biological perspective
- PMID: 23225628
- PMCID: PMC3588911
- DOI: 10.1002/pro.2201
Halogen bonding (X-bonding): a biological perspective
Abstract
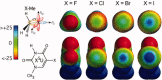
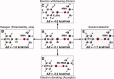
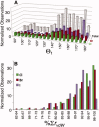
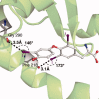
The concept of the halogen bond (or X-bond) has become recognized as contributing significantly to the specificity in recognition of a large class of halogenated compounds. The interaction is most easily understood as primarily an electrostatically driven molecular interaction, where an electropositive crown, or σ-hole, serves as a Lewis acid to attract a variety of electron-rich Lewis bases, in analogous fashion to a classic hydrogen bonding (H-bond) interaction. We present here a broad overview of X-bonds from the perspective of a biologist who may not be familiar with this recently rediscovered class of interactions and, consequently, may be interested in how they can be applied as a highly directional and specific component of the molecular toolbox. This overview includes a discussion for where X-bonds are found in biomolecular structures, and how their structure-energy relationships are studied experimentally and modeled computationally. In total, our understanding of these basic concepts will allow X-bonds to be incorporated into strategies for the rational design of new halogenated inhibitors against biomolecular targets or toward molecular engineering of new biological-based materials.
Copyright © 2012 The Protein Society.
Figures



References
-
- Guthries F. On the iodide of iodammonium. J Am Chem Soc. 1863;16:239–244.
-
- Hassel O. 17, 497–502: 1970. Scinece. - PubMed
-
- Lommerse JPM, Stone AJ, Taylor R, Allen FH. The nature and geometry of intramolecular interactions between halogens and oxygen or nitrogen. J Am Chem Soc. 1996;118:3108–3116.
-
- Legon AC. Prereactive complexes of dihalogens XY with Lewis bases B in the gas phase: a systematic case for the halogen analogue B···XY of the hydrogen bond B···HX. Angew Chem Int Ed Engl. 1999;38:2686–2714. - PubMed
-
- Metrangolo P, Neukirch H, Pilati T, Resnati G. Halogen bonding based recognition processes: a world parallel to hydrogen bonding. Acc Chem Res. 2005;38:386–395. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

