Clinical implementation of RNA signatures for pharmacogenomic decision-making
- PMID: 23226056
- PMCID: PMC3513222
- DOI: 10.2147/PGPM.S14888
Clinical implementation of RNA signatures for pharmacogenomic decision-making
Abstract
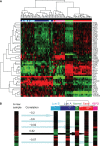
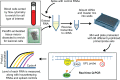
RNA profiling is increasingly used to predict drug response, dose, or toxicity based on analysis of drug pharmacokinetic or pharmacodynamic pathways. Before implementing multiplexed RNA arrays in clinical practice, validation studies are carried out to demonstrate sufficient evidence of analytic and clinical performance, and to establish an assay protocol with quality assurance measures. Pathologists assure quality by selecting input tissue and by interpreting results in the context of the input tissue as well as the technologies that were used and the clinical setting in which the test was ordered. A strength of RNA profiling is the array-based measurement of tens to thousands of RNAs at once, including redundant tests for critical analytes or pathways to promote confidence in test results. Instrument and reagent manufacturers are crucial for supplying reliable components of the test system. Strategies for quality assurance include careful attention to RNA preservation and quality checks at pertinent steps in the assay protocol, beginning with specimen collection and proceeding through the various phases of transport, processing, storage, analysis, interpretation, and reporting. Specimen quality is checked by probing housekeeping transcripts, while spiked and exogenous controls serve as a check on analytic performance of the test system. Software is required to manipulate abundant array data and present it for interpretation by a laboratory physician who reports results in a manner facilitating therapeutic decision-making. Maintenance of the assay requires periodic documentation of personnel competency and laboratory proficiency. These strategies are shepherding genomic arrays into clinical settings to provide added value to patients and to the larger health care system.
Keywords: RNA; clinical laboratory; microarray; preanalytic; quality assurance; translational.
Figures


Similar articles
-
Quality assurance of RNA expression profiling in clinical laboratories.J Mol Diagn. 2012 Jan;14(1):1-11. doi: 10.1016/j.jmoldx.2011.09.003. Epub 2011 Oct 20. J Mol Diagn. 2012. PMID: 22020152 Free PMC article. Review.
-
Implementation of point-of-care testing in a pediatric healthcare setting.Crit Rev Clin Lab Sci. 2019 Jun;56(4):239-246. doi: 10.1080/10408363.2019.1590306. Epub 2019 Apr 11. Crit Rev Clin Lab Sci. 2019. PMID: 30973797 Review.
-
Gene expression profiling for guiding adjuvant chemotherapy decisions in women with early breast cancer: an evidence-based and economic analysis.Ont Health Technol Assess Ser. 2010;10(23):1-57. Epub 2010 Dec 1. Ont Health Technol Assess Ser. 2010. PMID: 23074401 Free PMC article.
-
[Mycobacterial tests].Kekkaku. 2008 Jan;83(1):43-59. Kekkaku. 2008. PMID: 18283915 Japanese.
-
Computerized training and proficiency testing. International Academy of Cytology Task Force summary. Diagnostic Cytology Towards the 21st Century: An International Expert Conference and Tutorial.Acta Cytol. 1998 Jan-Feb;42(1):141-7. doi: 10.1159/000331539. Acta Cytol. 1998. PMID: 9479333 Review.
Cited by
-
Modulation of Small RNA Signatures by Astrocytes on Early Neurodegeneration Stages; Implications for Biomarker Discovery.Life (Basel). 2022 Oct 27;12(11):1720. doi: 10.3390/life12111720. Life (Basel). 2022. PMID: 36362875 Free PMC article. Review.
References
-
- Tumor Analysis Best Practices Working Group Expression profiling – best practices for data generation and interpretation in clinical trials. Nat Rev Genet. 2004;5(3):229–237. - PubMed
-
- Auer H, Newsom DL, Kornacker K. Expression profiling using Affymetrix GeneChip microarrays. Methods Mol Biol. 2009;509:35–46. - PubMed
-
- Fuscoe JC, Tong W, Shi L. QA/QC issues to aid regulatory acceptance of microarray gene expression data. Environ Mol Mutagen. 2007;48(5):349–353. - PubMed
-
- Hackett JL, Gutman SI. Introduction to the Food and Drug Administration (FDA) regulatory process. J Proteome Res. 2005;4(4):1110–1113. - PubMed
-
- Imbeaud S, Auffray C. The ‘39 steps’ in gene expression profiling: Critical issues and proposed best practices for microarray experiments. Drug Discov Today. 2005;10(17):1175–1182. - PubMed
LinkOut - more resources
Full Text Sources

