Microbial pathways in colonic sulfur metabolism and links with health and disease
- PMID: 23226130
- PMCID: PMC3508456
- DOI: 10.3389/fphys.2012.00448
Microbial pathways in colonic sulfur metabolism and links with health and disease
Abstract
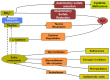
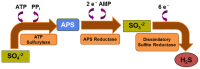
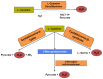
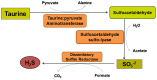
Sulfur is both crucial to life and a potential threat to health. While colonic sulfur metabolism mediated by eukaryotic cells is relatively well studied, much less is known about sulfur metabolism within gastrointestinal microbes. Sulfated compounds in the colon are either of inorganic (e.g., sulfates, sulfites) or organic (e.g., dietary amino acids and host mucins) origin. The most extensively studied of the microbes involved in colonic sulfur metabolism are the sulfate-reducing bacteria (SRB), which are common colonic inhabitants. Many other microbial pathways are likely to shape colonic sulfur metabolism as well as the composition and availability of sulfated compounds, and these interactions need to be examined in more detail. Hydrogen sulfide is the sulfur derivative that has attracted the most attention in the context of colonic health, and the extent to which it is detrimental or beneficial remains in debate. Several lines of evidence point to SRB or exogenous hydrogen sulfide as potential players in the etiology of intestinal disorders, inflammatory bowel diseases (IBDs) and colorectal cancer in particular. Generation of hydrogen sulfide via pathways other than dissimilatory sulfate reduction may be as, or more, important than those involving the SRB. We suggest here that a novel axis of research is to assess the effects of hydrogen sulfide in shaping colonic microbiome structure. Clearly, in-depth characterization of the microbial pathways involved in colonic sulfur metabolism is necessary for a better understanding of its contribution to colonic disorders and development of therapeutic strategies.
Keywords: colonic microbiota; colorectal cancer; hydrogen sulfide; inflammatory bowel disease; irritable bowel syndrome; sulfate-reducing bacteria; sulfur.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

