Expression of Caytaxin protein in Cayman Ataxia mouse models correlates with phenotype severity
- PMID: 23226316
- PMCID: PMC3511541
- DOI: 10.1371/journal.pone.0050570
Expression of Caytaxin protein in Cayman Ataxia mouse models correlates with phenotype severity
Abstract
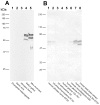
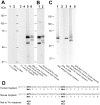
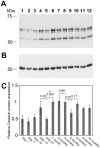
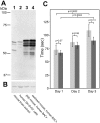
Caytaxin is a highly-conserved protein, which is encoded by the Atcay/ATCAY gene. Mutations in Atcay/ATCAY have been identified as causative of cerebellar disorders such as the rare hereditary disease Cayman ataxia in humans, generalized dystonia in the dystonic (dt) rat, and marked motor defects in three ataxic mouse lines. While several lines of evidence suggest that Caytaxin plays a critical role in maintaining nervous system processes, the physiological function of Caytaxin has not been fully characterized. In the study presented here, we generated novel specific monoclonal antibodies against full-length Caytaxin to examine endogenous Caytaxin expression in wild type and Atcay mutant mouse lines. Caytaxin protein is absent from brain tissues in the two severely ataxic Atcay(jit) (jittery) and Atcay(swd) (sidewinder) mutant lines, and markedly decreased in the mildly ataxic/dystonic Atcay(ji-hes) (hesitant) line, indicating a correlation between Caytaxin expression and disease severity. As the expression of wild type human Caytaxin in mutant sidewinder and jittery mice rescues the ataxic phenotype, Caytaxin's physiological function appears to be conserved between the human and mouse orthologs. Across multiple species and in several neuronal cell lines Caytaxin is expressed as several protein isoforms, the two largest of which are caused by the usage of conserved methionine translation start sites. The work described in this manuscript presents an initial characterization of the Caytaxin protein and its expression in wild type and several mutant mouse models. Utilizing these animal models of human Cayman Ataxia will now allow an in-depth analysis to elucidate Caytaxin's role in maintaining normal neuronal function.
Conflict of interest statement
Figures

Similar articles
-
Genome-Wide Association Study and Subsequent Exclusion of ATCAY as a Candidate Gene Involved in Equine Neuroaxonal Dystrophy Using Two Animal Models.Genes (Basel). 2020 Jan 10;11(1):82. doi: 10.3390/genes11010082. Genes (Basel). 2020. PMID: 31936863 Free PMC article.
-
Mutations in a novel gene encoding a CRAL-TRIO domain cause human Cayman ataxia and ataxia/dystonia in the jittery mouse.Nat Genet. 2003 Nov;35(3):264-9. doi: 10.1038/ng1255. Epub 2003 Oct 12. Nat Genet. 2003. PMID: 14556008
-
Expression and localization of Cayman ataxia-related protein, Caytaxin, is regulated in a developmental- and spatial-dependent manner.Brain Res. 2007 Jan 19;1129(1):100-9. doi: 10.1016/j.brainres.2006.10.068. Epub 2006 Dec 8. Brain Res. 2007. PMID: 17157273
-
PTPRR protein tyrosine phosphatase isoforms and locomotion of vesicles and mice.Cerebellum. 2009 Jun;8(2):80-8. doi: 10.1007/s12311-008-0088-y. Epub 2009 Jan 10. Cerebellum. 2009. PMID: 19137382 Free PMC article. Review.
-
Mutant mice as a model for cerebellar ataxia.Prog Neurobiol. 2001 Apr;63(5):489-540. doi: 10.1016/s0301-0082(00)00024-1. Prog Neurobiol. 2001. PMID: 11164620 Review.
Cited by
-
Alterations in cerebellar physiology are associated with a stiff-legged gait in Atcay(ji-hes) mice.Neurobiol Dis. 2014 Jul;67:140-8. doi: 10.1016/j.nbd.2014.03.020. Epub 2014 Apr 12. Neurobiol Dis. 2014. PMID: 24727095 Free PMC article.
-
Distinct DNA-based epigenetic switches trigger transcriptional activation of silent genes in human dermal fibroblasts.Sci Rep. 2014 Jan 24;4:3843. doi: 10.1038/srep03843. Sci Rep. 2014. PMID: 24457603 Free PMC article.
-
Degradation of Caytaxin Causes Learning and Memory Deficits via Activation of DAPK1 in Aging.Mol Neurobiol. 2019 May;56(5):3368-3379. doi: 10.1007/s12035-018-1312-5. Epub 2018 Aug 17. Mol Neurobiol. 2019. PMID: 30120735
-
Genome-Wide Association Study and Subsequent Exclusion of ATCAY as a Candidate Gene Involved in Equine Neuroaxonal Dystrophy Using Two Animal Models.Genes (Basel). 2020 Jan 10;11(1):82. doi: 10.3390/genes11010082. Genes (Basel). 2020. PMID: 31936863 Free PMC article.
-
Vocal development in dystonic rats.Physiol Rep. 2015 Apr;3(4):e12350. doi: 10.14814/phy2.12350. Physiol Rep. 2015. PMID: 25907786 Free PMC article.
References
-
- Bomar JM, Benke PJ, Slattery EL, Puttagunta R, Taylor LP, et al. (2003) Mutations in a novel gene encoding a CRAL-TRIO domain cause human Cayman ataxia and ataxia/dystonia in the jittery mouse. Nat Genet 35: 264–269. - PubMed
-
- Kumar A.K MJaSC (2004) Genetic Disorders in the Cayman Islands. Cayman Islands Country Conference.
-
- DeOme K (1945) A new recessive lethal mutation in mice. University of California publications in zoology 53: 41–65.
-
- Kapfhamer D, Sweet HO, Sufalko D, Warren S, Johnson KR, et al. (1996) The neurological mouse mutations jittery and hesitant are allelic and map to the region of mouse chromosome 10 homologous to 19p13.3. Genomics 35: 533–538. - PubMed
-
- Xiao J, Ledoux MS (2005) Caytaxin deficiency causes generalized dystonia in rats. Brain Res Mol Brain Res 141: 181–192. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

