Haemophilus influenzae OxyR: characterization of its regulation, regulon and role in fitness
- PMID: 23226321
- PMCID: PMC3511568
- DOI: 10.1371/journal.pone.0050588
Haemophilus influenzae OxyR: characterization of its regulation, regulon and role in fitness
Abstract
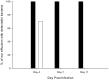
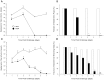
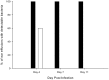
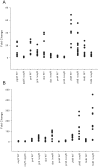
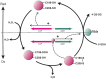
To prevent damage by reactive oxygen species, many bacteria have evolved rapid detection and response systems, including the OxyR regulon. The OxyR system detects reactive oxygen and coordinates the expression of numerous defensive antioxidants. In many bacterial species the coordinated OxyR-regulated response is crucial for in vivo survival. Regulation of the OxyR regulon of Haemophilus influenzae was examined in vitro, and significant variation in the regulated genes of the OxyR regulon among strains of H. influenzae was observed. Quantitative PCR studies demonstrated a role for the OxyR-regulated peroxiredoxin/glutaredoxin as a mediator of the OxyR response, and also indicated OxyR self-regulation through a negative feedback loop. Analysis of transcript levels in H. influenzae samples derived from an animal model of otitis media demonstrated that the members of the OxyR regulon were actively upregulated within the chinchilla middle ear. H. influenzae mutants lacking the oxyR gene exhibited increased sensitivity to challenge with various peroxides. The impact of mutations in oxyR was assessed in various animal models of H. influenzae disease. In paired comparisons with the corresponding wild-type strains, the oxyR mutants were unaffected in both the chinchilla model of otitis media and an infant model of bacteremia. However, in weanling rats the oxyR mutant was significantly impaired compared to the wild-type strain. In contrast, in all three animal models when infected with a mixture of equal numbers of both wild-type and mutant strains the mutant strain was significantly out competed by the wild-type strain. These findings clearly establish a crucial role for OxyR in bacterial fitness.
Conflict of interest statement
Figures

References
-
- Turk DC (1984) The pathogenicity of Haemophilus influenzae . J Med Microbiol 18: 1–16. - PubMed
-
- Murphy TF, Faden H, Bakaletz LO, Kyd JM, Forsgren A, et al. (2009) Nontypeable Haemophilus influenzae as a pathogen in children. Pediatr Infect Dis J 28: 43–48. - PubMed
-
- Morton DJ, Stull TL (2004) Haemophilus. In: Crosa JH, Mey AR, Payne SM, editors. Iron Transport in Bacteria. Washington, DC: American Society for Microbiology. 273–292.
-
- Morton DJ, Smith A, Ren Z, Madore LL, VanWagoner TM, et al. (2004) Identification of a haem-utilization protein (Hup) in Haemophilus influenzae . Microbiology 150: 3923–3933. - PubMed
-
- Morton DJ, Madore LL, Smith A, VanWagoner TM, Seale TW, et al. (2005) The heme-binding lipoprotein (HbpA) of Haemophilus influenzae: role in heme utilization. FEMS Microbiol Lett 253: 193–199. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

