Functional characterization of two M42 aminopeptidases erroneously annotated as cellulases
- PMID: 23226342
- PMCID: PMC3511314
- DOI: 10.1371/journal.pone.0050639
Functional characterization of two M42 aminopeptidases erroneously annotated as cellulases
Abstract
Several aminopeptidases of the M42 family have been described as tetrahedral-shaped dodecameric (TET) aminopeptidases. A current hypothesis suggests that these enzymes are involved, along with the tricorn peptidase, in degrading peptides produced by the proteasome. Yet the M42 family remains ill defined, as some members have been annotated as cellulases because of their homology with CelM, formerly described as an endoglucanase of Clostridium thermocellum. Here we describe the catalytic functions and substrate profiles CelM and of TmPep1050, the latter having been annotated as an endoglucanase of Thermotoga maritima. Both enzymes were shown to catalyze hydrolysis of nonpolar aliphatic L-amino acid-pNA substrates, the L-leucine derivative appearing as the best substrate. No significant endoglucanase activity was measured, either for TmPep1050 or CelM. Addition of cobalt ions enhanced the activity of both enzymes significantly, while both the chelating agent EDTA and bestatin, a specific inhibitor of metalloaminopeptidases, proved inhibitory. Our results strongly suggest that one should avoid annotating members of the M42 aminopeptidase family as cellulases. In an updated assessment of the distribution of M42 aminopeptidases, we found TET aminopeptidases to be distributed widely amongst archaea and bacteria. We additionally observed that several phyla lack both TET and tricorn. This suggests that other complexes may act downstream from the proteasome.
Conflict of interest statement
Figures

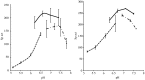
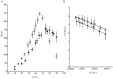
References
-
- Groll M, Bochtler M, Brandstetter H, Clausen T, Huber R (2005) Molecular machines for protein degradation. ChemBioChem 6: 222–256 doi:10.1002/cbic.200400313. - DOI - PubMed
-
- Jenal U, Hengge-Aronis R (2003) Regulation by proteolysis in bacterial cells. Curr Opin Microbiol 6: 163–172. - PubMed
-
- Pruteanu M, Baker TA (2009) Proteolysis in the SOS response and metal homeostasis in Escherichia coli . Res Microbiol 160: 677–683 doi:10.1016/j.resmic.2009.08.012. - DOI - PMC - PubMed
-
- Dahlmann B, Kopp F, Kuehn L, Niedel B, Pfeifer G, et al. (1989) The multicatalytic proteinase (prosome) is ubiquitous from eukaryotes to archaebacteria. FEBS Lett 251: 125–131. - PubMed
-
- Tamura T, Nagy I, Lupas A, Lottspeich F, Cejka Z, et al. (1995) The first characterization of a eubacterial proteasome: the 20S complex of Rhodococcus . Curr Biol 5: 766–774. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

