Identification and testing of reference genes for Sesame gene expression analysis by quantitative real-time PCR
- PMID: 23229061
- PMCID: PMC3579469
- DOI: 10.1007/s00425-012-1805-9
Identification and testing of reference genes for Sesame gene expression analysis by quantitative real-time PCR
Abstract
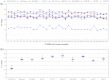
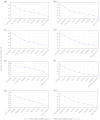
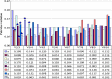
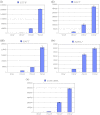
Sesame (Sesamum indicum L.) is an ancient and important oilseed crop. However, few sesame reference genes have been selected for quantitative real-time PCR until now. Screening and validating reference genes is a requisite for gene expression normalization in sesame functional genomics research. In this study, ten candidate reference genes, i.e., SiACT, SiUBQ6, SiTUB, Si18S rRNA, SiEF1α, SiCYP, SiHistone, SiDNAJ, SiAPT and SiGAPDH, were chosen and examined systematically in 32 sesame samples. Three qRT-PCR analysis methods, i.e., geNorm, NormFinder and BestKeeper, were evaluated systematically. Results indicated that all ten candidate reference genes could be used as reference genes in sesame. SiUBQ6 and SiAPT were the optimal reference genes for sesame plant development; SiTUB was suitable for sesame vegetative tissue development, SiDNAJ for pathogen treatment, SiHistone for abiotic stress, SiUBQ6 for bud development and SiACT for seed germination. As for hormone treatment and seed development, SiHistone, SiCYP, SiDNAJ or SiUBQ6, as well as SiACT, SiDNAJ, SiTUB or SiAPT, could be used as reference gene, respectively. To illustrate the suitability of these reference genes, we analyzed the expression variation of three functional sesame genes of SiSS, SiLEA and SiGH in different organs using the optimal qRT-PCR system for the first time. The stability levels of optimal and worst reference genes screened for seed development, anther sterility and plant development were validated in the qRT-PCR normalization. Our results provided a reference gene application guideline for sesame gene expression characterization using qRT-PCR system.
Figures




Similar articles
-
Selection of Reliable Reference Genes for Gene Expression Studies of a Promising Oilseed Crop, Plukenetia volubilis, by Real-Time Quantitative PCR.Int J Mol Sci. 2015 Jun 3;16(6):12513-30. doi: 10.3390/ijms160612513. Int J Mol Sci. 2015. PMID: 26047338 Free PMC article.
-
Identification and evaluation of new reference genes in Gossypium hirsutum for accurate normalization of real-time quantitative RT-PCR data.BMC Plant Biol. 2010 Mar 21;10:49. doi: 10.1186/1471-2229-10-49. BMC Plant Biol. 2010. PMID: 20302670 Free PMC article.
-
Identification and validation of reference genes for Populus euphratica gene expression analysis during abiotic stresses by quantitative real-time PCR.Physiol Plant. 2014 Nov;152(3):529-45. doi: 10.1111/ppl.12206. Epub 2014 Jun 2. Physiol Plant. 2014. PMID: 24720378
-
Genome-wide association study and its applications in the non-model crop Sesamum indicum.BMC Plant Biol. 2021 Jun 22;21(1):283. doi: 10.1186/s12870-021-03046-x. BMC Plant Biol. 2021. PMID: 34157965 Free PMC article. Review.
-
Omics technologies towards sesame improvement: a review.Mol Biol Rep. 2023 Aug;50(8):6885-6899. doi: 10.1007/s11033-023-08551-w. Epub 2023 Jun 16. Mol Biol Rep. 2023. PMID: 37326753 Review.
Cited by
-
Identification and validation of Aeluropus littoralis reference genes for Quantitative Real-Time PCR Normalization.J Biol Res (Thessalon). 2016 Jul 19;23:18. doi: 10.1186/s40709-016-0053-8. eCollection 2016 Dec. J Biol Res (Thessalon). 2016. PMID: 27437194 Free PMC article.
-
Selection and validation of reference genes for normalization of qRT-PCR gene expression in wheat (Triticum durum L.) under drought and salt stresses.J Genet. 2018 Dec;97(5):1433-1444. J Genet. 2018. PMID: 30555091
-
Validation of reference genes for quantitative RT-PCR normalization in Suaeda aralocaspica, an annual halophyte with heteromorphism and C4 pathway without Kranz anatomy.PeerJ. 2016 Feb 11;4:e1697. doi: 10.7717/peerj.1697. eCollection 2016. PeerJ. 2016. PMID: 26893974 Free PMC article.
-
Reference gene screening for analyzing gene expression in the heart, liver, spleen, lung and kidney of forest musk deer.J Vet Med Sci. 2021 Nov 16;83(11):1750-1759. doi: 10.1292/jvms.21-0281. Epub 2021 Oct 7. J Vet Med Sci. 2021. PMID: 34615843 Free PMC article.
-
Identification of a Sidwf1 gene controlling short internode length trait in the sesame dwarf mutant dw607.Theor Appl Genet. 2020 Jan;133(1):73-86. doi: 10.1007/s00122-019-03441-x. Epub 2019 Nov 5. Theor Appl Genet. 2020. PMID: 31686114
References
-
- Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. - DOI - PubMed
-
- Bateer S, Qiang L, Hui Z, Agula H, Xiaoyun J, Fengyun G. Study on mRNA differential expression in male sterile and fertile flower buds of dominant genomic male sterile flax and sequence analysis of differential fragments. Biotechnol Bull. 2009;8:67–70.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

