RNA helicases in splicing
- PMID: 23229095
- PMCID: PMC3590240
- DOI: 10.4161/rna.22547
RNA helicases in splicing
Abstract
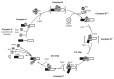
In eukaryotic cells, introns are spliced from pre-mRNAs by the spliceosome. Both the composition and the structure of the spliceosome are highly dynamic, and eight DExD/H RNA helicases play essential roles in controlling conformational rearrangements. There is evidence that the various helicases are functionally and physically connected with each other and with many other factors in the spliceosome. Understanding the dynamics of those interactions is essential to comprehend the mechanism and regulation of normal as well as of pathological splicing. This review focuses on recent advances in the characterization of the splicing helicases and their interactions, and highlights the deep integration of splicing helicases in global mRNP biogenesis pathways.
Keywords: ATPase; DEAD-box; DEAH-box; DExD/H helicase; protein partners.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
