Molecular diagnosis to discriminate pathogen and apathogen species of the hybrid Verticillium longisporum on the oilseed crop Brassica napus
- PMID: 23229565
- PMCID: PMC3647090
- DOI: 10.1007/s00253-012-4530-1
Molecular diagnosis to discriminate pathogen and apathogen species of the hybrid Verticillium longisporum on the oilseed crop Brassica napus
Abstract
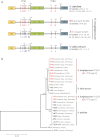
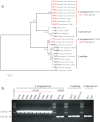
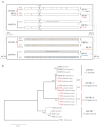
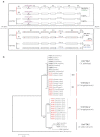
The cruciferous fungal pathogen Verticillium longisporum represents an allodiploid hybrid with long spores and almost double the amount of nuclear DNA compared to other Verticillium species. V. longisporum evolved at least three times by hybridization. In Europe, virulent A1xD1 and avirulent A1xD3 hybrids were isolated from the oilseed crop Brassica napus. Parental A1 or D1 species are yet unknown whereas the D3 lineage represents Verticillium dahliae. Eleven V. longisporum isolates from Europe or California corresponding to hybrids A1xD1 or A1xD3 were compared. A single characteristic type of nuclear ribosomal DNA could be assigned to each hybrid lineage. The two avirulent A1xD3 isolates carried exclusively D3 ribosomal DNA (rDNA) which corresponds to V. dahliae. The rDNA of all nine A1xD1 isolates is identical but distinct from D3 and presumably originates from A1. Both hybrid lineages carry two distinct isogene pairs of four conserved regulatory genes corresponding to either A1 or D1/D3. D1 and D3 paralogues differ in several single nucleotide polymorphisms. Southern hybridization patterns confirmed differences between the A1 and D1/D3 isogenes and resulted in similar patterns for D1 and D3. Distinct signatures of the Verticillium transcription activator (VTA)2 regulatory isogene pair allow identification of V. longisporum hybrids by a single polymerase chain reaction and the separation from haploid species as V. dahliae or Verticillium albo-atrum. The combination between VTA2 signature and rDNA type identification represents an attractive diagnostic tool to discriminate allodiploid from haploid Verticillia and to distinguish between A1xD1 and A1xD3 hybrids which differ in their virulence towards B. napus.
Figures


References
-
- Agrios GN. Plant pathology. Burlington, USA: Elsevier; 2005.
-
- Bayram O, Krappmann S, Ni M, Bok JW, Helmstaedt K, Valerius O, Braus-Stromeyer S, Kwon NJ, Keller NP, Yu JH, Braus GH (2008) VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320:1504–1506 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

