Acute B lymphoblastic leukaemia-propagating cells are present at high frequency in diverse lymphoblast populations
- PMID: 23229821
- PMCID: PMC3569652
- DOI: 10.1002/emmm.201201703
Acute B lymphoblastic leukaemia-propagating cells are present at high frequency in diverse lymphoblast populations
Abstract
Leukaemia-propagating cells are more frequent in high-risk acute B lymphoblastic leukaemia than in many malignancies that follow a hierarchical cancer stem cell model. It is unclear whether this characteristic can be more universally applied to patients from non-'high-risk' sub-groups and across a broad range of cellular immunophenotypes. Here, we demonstrate in a wide range of primary patient samples and patient samples previously passaged through mice that leukaemia-propagating cells are found in all populations defined by high or low expression of the lymphoid differentiation markers CD10, CD20 or CD34. The frequency of leukaemia-propagating cells and their engraftment kinetics do not differ between these populations. Transcriptomic analysis of CD34(high) and CD34(low) blasts establishes their difference and their similarity to comparable normal progenitors at different stages of B-cell development. However, consistent with the functional similarity of these populations, expression signatures characteristic of leukaemia propagating cells in acute myeloid leukaemia fail to distinguish between the different populations. Together, these findings suggest that there is no stem cell hierarchy in acute B lymphoblastic leukaemia.
Copyright © 2013 The Authors. Published by John Wiley and Sons, Ltd on behalf of EMBO.
Figures
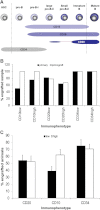
Expression of the cell surface antigens CD10, CD19, CD20 and CD34 during normal human B-cell development (van Zelm et al, 2005).
Percentage of engrafting primary patient and primograft samples for the different populations (CD10low/high, CD20low/high, CD34low/high). Black bars provide the data for primary and white bars for primograft samples.
Weighted mean of engraftment in mice transplanted with at least 1000 blasts sorted for CD10, CD20 or CD34 expression. The analysis takes into account that different numbers of mice were transplanted with cells from different samples. For data of engraftment of mice transplanted with cells sorted for expression of CD10 and CD20, see also Supporting Information Tables S1 and S2. The CD34 data are taken from the limiting dilutions experiments (Table 2) and include only the mice transplanted with 1000 cells.

Engraftment of primografted CD10low and CD10high cells (patient L4951).
Engraftment of primografted CD20low and CD20high cells (patient EMCR1).
Engraftment of primografted CD34low and CD34high cells (patient EMCR1).
Engraftment of primary CD34low and CD34high cells (patient L784).

Expression of TERT in CD34high and CD34low umbilical cord blood cells and leukaemic blasts. Umbilical cord blood shows higher expression of TERT in immature CD34high cells whilst no difference is seen between immature CD34high and CD34low blasts. TERT expression in blasts is comparable with, or higher than, that in CD34high umbilical cord blood progenitor cells. Expression values are 2-ΔCt using GAPDH as the reference gene. For purity of cells sorted for CD34 expression, see Supporting Information Fig S6.
The PCA using a haematopoietic “self-renewal” signature containing 59 genes (213 probes) (Kim et al, 2009) also failed to separate CD34high and CD34low B-ALL populations (Supporting Information Fig S4). Here, we show the heatmap of the expression of 41 selected probes from this “self-renewal” signature.
PCA plot generated using an AML “stem cell” signature (Eppert et al, 2011) that separates Lin-CD34highCD38low blasts (dark purple symbols) from more mature CD34low (pink symbols) blasts in AML (Gentles et al, 2010) but fails to detect any significant differences in “self-renewal” gene expression amongst the leukaemic subpopulations (black & white symbols) in B-ALL.

A,B. Engraftment kinetics of CD10high and CD10low cells after transplantation of 100 (n = 4) or 1000–2000 blasts (n ≥ 4).
C,D. Engraftment kinetics of CD20high and CD20low cells after transplantation of 100–300 (n ≥ 8) or 500–3000 blasts (n ≥ 21).
E,F. Engraftment kinetics of CD34high and CD34low cells after transplantation of 100 (n ≥ 6, last points n = 2 for CD34high and n = 3 for CD34low) or 1000 blasts (n ≥ 9).

In normal haematopoietic stem cells and AML, self-renewal is tightly regulated by a differentiation-dependent loss of self-renewal.
In B-ALL, clonal expansion is tightly regulated by the dependence of positive survival and proliferation signals. Without those lymphoid blasts undergo apoptosis.
Comment in
-
The cancer stem cell model: B cell acute lymphoblastic leukaemia breaks the mould.EMBO Mol Med. 2013 Jan;5(1):7-9. doi: 10.1002/emmm.201202207. Epub 2012 Dec 11. EMBO Mol Med. 2013. PMID: 23229878 Free PMC article. No abstract available.
References
-
- Anderson K, Lutz C, van Delft FW, Bateman CM, Guo Y, Colman SM, Kempski H, Moorman AV, Titley I, Swansbury J, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469:356–361. - PubMed
-
- Andersson A, Ritz C, Lindgren D, Eden P, Lassen C, Heldrup J, Olofsson T, Rade J, Fontes M, Porwit-Macdonald A, et al. Microarray-based classification of a consecutive series of 121 childhood acute leukemias: prediction of leukemic and genetic subtype as well as of minimal residual disease status. Leukemia. 2007;21:1198–1203. - PubMed
-
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

