Novel cholix toxin variants, ADP-ribosylating toxins in Vibrio cholerae non-O1/non-O139 strains, and their pathogenicity
- PMID: 23230295
- PMCID: PMC3553811
- DOI: 10.1128/IAI.00982-12
Novel cholix toxin variants, ADP-ribosylating toxins in Vibrio cholerae non-O1/non-O139 strains, and their pathogenicity
Abstract
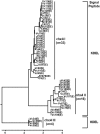
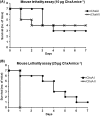
Cholix toxin (ChxA) is a recently discovered exotoxin in Vibrio cholerae which has been characterized as a third member of the eukaryotic elongation factor 2-specific ADP-ribosyltransferase toxins, in addition to exotoxin A of Pseudomonas aeruginosa and diphtheria toxin of Corynebacterium diphtheriae. These toxins consist of three characteristic domains for receptor binding, translocation, and catalysis. However, there is little information about the prevalence of chxA and its genetic variations and pathogenic mechanisms. In this study, we screened the chxA gene in a large number (n = 765) of V. cholerae strains and observed its presence exclusively in non-O1/non-O139 strains (27.0%; 53 of 196) and not in O1 (n = 485) or O139 (n = 84). Sequencing of these 53 chxA genes generated 29 subtypes which were grouped into three clusters designated chxA I, chxA II, and chxA III. chxA I belongs to the prototype, while chxA II and chxA III are newly discovered variants. ChxA II and ChxA III had unique receptor binding and catalytic domains, respectively, in comparison to ChxA I. Recombinant ChxA I (rChxA I) and rChxA II but not rChxA III showed variable cytotoxic effects on different eukaryotic cells. Although rChxA II was more lethal to mice than rChxA I when injected intravenously, no enterotoxicity of any rChxA was observed in a rabbit ileal loop test. Hepatocytes showed coagulation necrosis in rChxA I- or rChxA II-treated mice, seemingly the major target for ChxA. The present study illustrates the potential of ChxA as an important virulence factor in non-O1/non-O139 V. cholerae, which may be associated with extraintestinal infections rather than enterotoxicity.
Figures




References
-
- WHO July 2012. Cholera. Media centre fact sheet no. 107. WHO, Geneva, Switzerland: http://www.who.int/mediacentre/factsheets/fs107/en/.
-
- Sharma C, Thungapathra M, Ghosh A, Mukhopadhyay AK, Basu A, Mitra R, Basu I, Bhattacharya SK, Shimada T, Ramamurthy T, Takeda T, Yamasaki S, Takeda Y, Nair GB. 1998. Molecular analysis of Vibrio cholerae non-O1, non-O139 associated with an unusual upsurge in the incidence of cholera-like disease in Calcutta, India. J. Clin. Microbiol. 36:756–763 - PMC - PubMed
-
- Chatterjee S, Ghosh K, Raychoudhuri A, Chowdhury G, Bhattacharya MK, Mukhopadhyay AK, Ramamurthy T, Bhattacharya SK, Klose KE, Nandy RK. 2009. Incidence, virulence factors, and clonality among clinical strains of non-O1, non-O139 Vibrio cholerae isolates from hospitalized diarrheal patients in Kolkata, India. J. Clin. Microbiol. 47:1087–1095 - PMC - PubMed
-
- Dziejman M, Serruto D, Tam VC, Sturtevant D, Diraphat P, Faruque SM, Rahman MH, Heidelberg JF, Decker J, Li L, Montgomery KT, Grills G, Kucherlapati R, Mekalanos JJ. 2005. Genomic characterization of non-O1, non-O139 Vibrio cholerae reveals genes for a type III secretion system. Proc. Natl. Acad. Sci. U. S. A. 102:3465–3470 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

